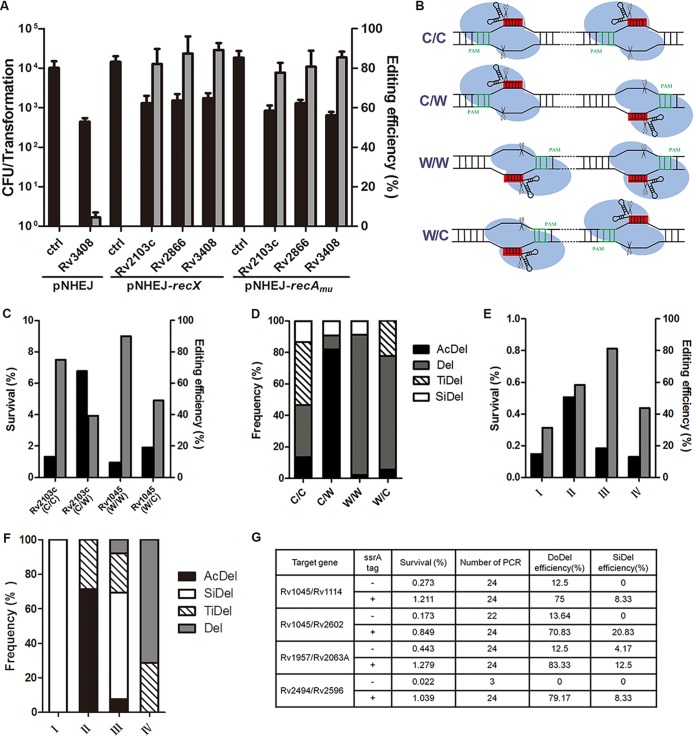
FIG 5.
CRISPR-NHEJ-assisted genome editing in M. tuberculosis. (A) CRISPR-Cas9Sth1 cleavage combined with NHEJ repair leads to efficient genome editing in M. tuberculosis H37Ra. sgRNA-expressing plasmids were electroporated into M. tuberculosis cells harboring various NHEJ-expressing plasmids. Transformation efficiency was defined as the total number of CFU obtained per transformation, and editing efficiency was calculated as the ratio of the number of edited events to the total number of colonies tested. Bars represent mean values ± standard deviations from two independent experiments. (B to D) Analysis of NHEJ efficiencies induced by paired Cas9Sth1-sgRNAs in four different orientations. Four different Cas9Sth1-sgRNA orientations were guided by paired PAMs. C/C, C/W, W/W, and W/C orientations were defined by the positioning of the paired PAMs on either the Watson strand (W) or the Crick strand (C). (E, F) Analysis of editing efficiency induced by paired Cas9Sth1-sgRNAs at the RD1 regions (I, II, III, and IV represent four different Cas9Sth1-sgRNA orientations guided by paired PAMs; see Fig. S2B). (G) Simultaneous generation of double mutations in M. tuberculosis using paired sgRNAs with Cas9Sth1 or Cas9Sth1-ssrA. Survival was defined as the ratio of the number of CFU obtained from the indicated sgRNA to the number of CFU obtained from the control plasmids, and editing efficiency was calculated as the ratio of the number of edited events to the total number of colonies tested; at least 32 colonies were analyzed by PCR and sequenced for each group. DoDel, double deletion. (D, F) The frequencies of accurate deletion (AcDel), single deletion (SiDel), two individual deletions (TiDel), and deletion (Del) were calculated as the ratios of the number of events from each group to the total number of edited events.