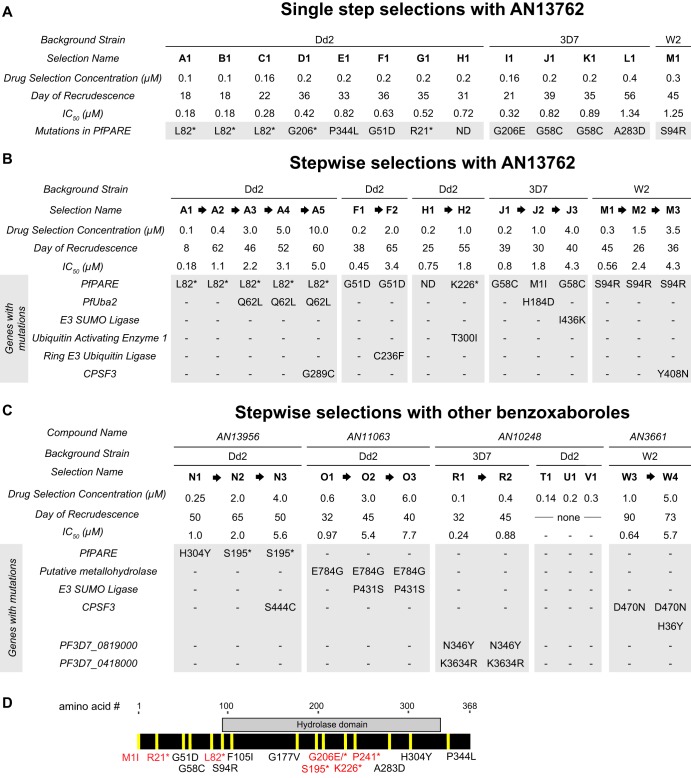
FIG 2.
Schematics showing resistance selections and SNPs identified in selected parasites (shaded). Stop codons are indicated by asterisks. (A) Single-step selections with AN13762 for indicated strains. We observed no dominant mutation (>60% mutant reads at position) in strains D1, E1, F1, and G1. Upon closer examination, several low-prevalence (20 to 50%) mutations were identified in PfPARE, with the most abundant mutation noted (Data Set S1). (B) Stepwise selection with AN13762. In strain H2, a single nucleotide insertion (435529_435530) and subsequent frameshift leading to a premature stop codon was observed. (C) Stepwise selection with other benzoxaboroles. For selections T1, U1, and V1, parasites did not recrudesce over 90 days. The results for AN3661-resistant strains W3 and W4 were published previously (Sonoiki et al. [15]). (D) Map of nonsynonymous mutations selected in PfPARE.