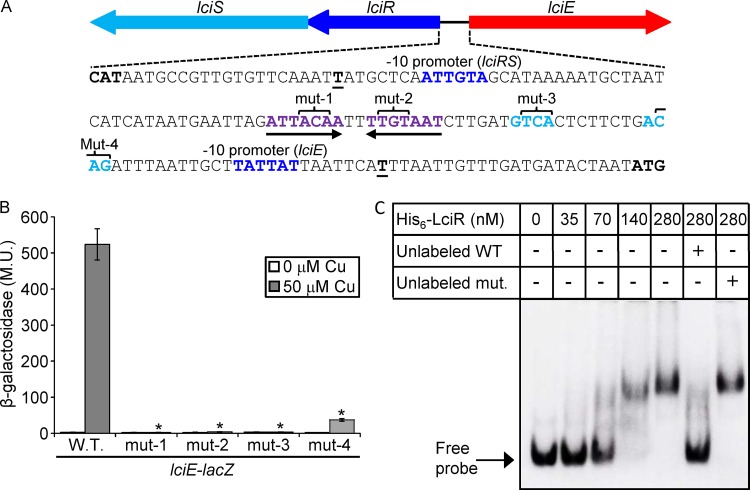
FIG 4.
LciR directly regulates the expression of lciE using a conserved regulatory element. (A) The intergenic DNA sequence located between lciR and lciE. The lciR and lciE −10 promoter elements are in dark blue, and the nucleotides representing the putative LciR consensus are in purple (the inverted-repeat sequence) or light blue (the two sequences located between the inverted repeat and the lciE −10 promoter); the inverted repeat is also marked with arrows. The transcription start sites are boldface and underlined. The four triplets of base pairs mutated in each part of the suspected regulatory element are indicated. (B) The effect of mutations in the putative LciR regulatory element. The levels of expression of wild-type lciE-lacZ fusion and the lciE-lacZ fusions containing mutations in the nucleotides marked in panel A were examined with and without 50 μM copper. The levels of expression of the lacZ fusions were found to be significantly different (*, P < 10−5, paired Student's t test) between the fusions containing the wild-type regulatory region and the mutated regulatory region under the same copper concentrations. β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results from at least three different experiments. (C) L. pneumophila His6-LciR protein binds to the lciE regulatory region. Gel mobility shift assay was performed with purified His6-LciR protein and the DIG-labeled lciE regulatory region. The first lane did not contain any protein. The rest of the lanes contained increasing amounts of the His6-LciR protein in 2-fold increments, starting from 35 nM. Competition was performed using unlabeled probe as a specific competitor (unlabeled WT) or a probe containing a mutation in the LciR regulatory element (unlabeled mut.).