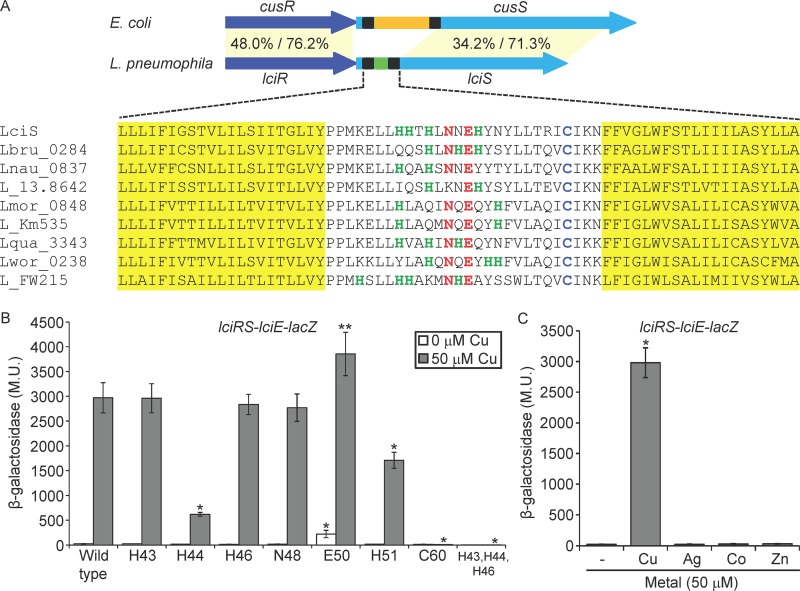
FIG 5.
Identification of amino acids required for copper induction located in the LciS periplasmic sensor domain. (A) Schematic illustration of the L. pneumophila lciRS and the E. coli cusRS genes. The periplasmic domain of the nine Legionella SHKs is much smaller than the periplasmic domain found in other bacteria, such as E. coli (28 amino acids compared to about 150 amino acids, respectively). In addition, protein sequence alignment of the LciS transmembrane regions and periplasmic domains from nine Legionella species harboring the LciRS TCS is shown. The amino acids that compose the SHK predicted transmembrane domains are colored yellow. Amino acids found in the periplasmic domain which might be involved in copper-binding are marked by different colors: histidine residues, green; cysteine, light blue; and asparagine and glutamic acid, red. (B) The effect of mutations in the periplasmic domain of the L. pneumophila LciS on copper induction. The levels of expression of wild-type lciRS-lciE-lacZ fusion and the lciRS-lciE-lacZ fusions containing mutations in the amino acids indicated below the bars, with and without 50 μM copper. The levels of expression of the lacZ fusions were found to be significantly different (**, P < 10−4; *, P < 10−5; both by paired Student's t test) between the mutated fusions and the wild-type fusion with or without copper. (C) The effect of different metals on lciE expression. The levels of expression of wild-type lciRS-lciE-lacZ fusion after exposure to the metals indicated below the bars (50 μM). The level of expression of the lacZ fusion was found to be significantly different (P < 10−5, paired Student's t test) between expression with metals and the one without metal exposure. β-Galactosidase activity was measured as described in Materials and Methods. Data (expressed in Miller units [M.U.]) are the averages ± standard deviations (error bars) of the results from at least three different experiments.