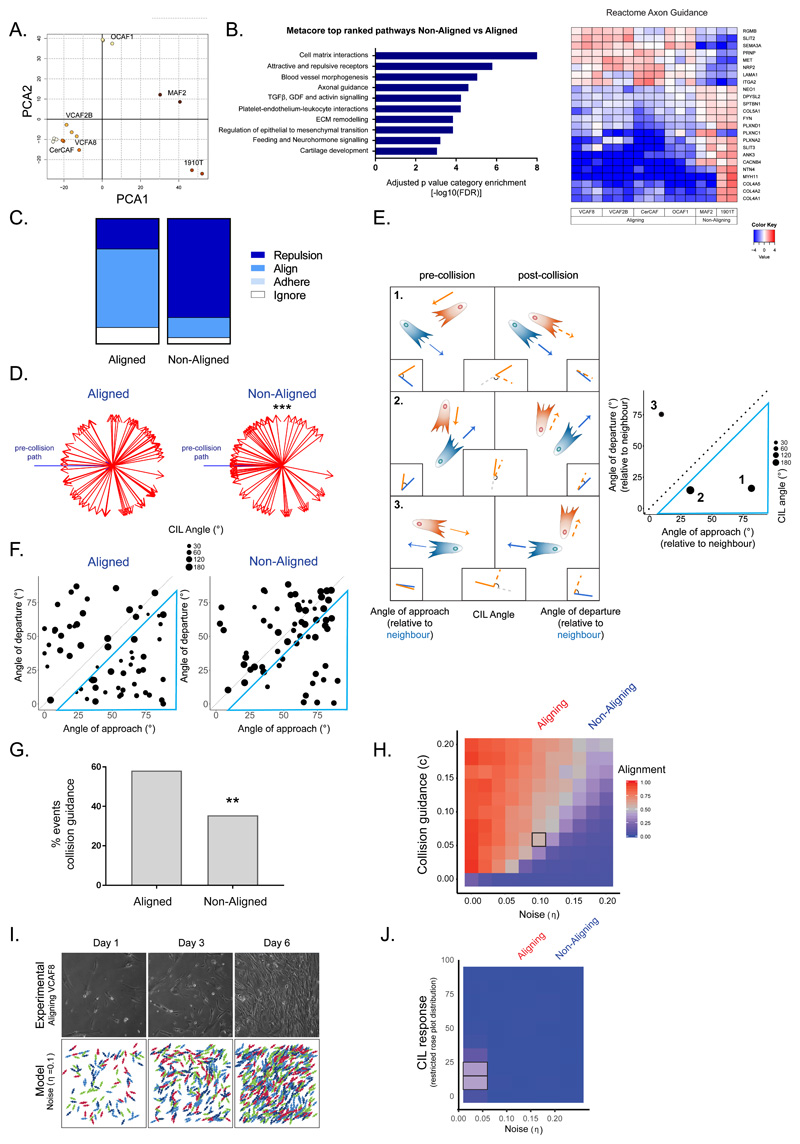
Figure 3. Aligned fibroblasts show supressed contact inhibition of locomotion and elevated collision guidance.
A. Principal component analysis showing segregation of fibroblast RNAseq data (n=16 from 3 independent experiments). B. Top differentially regulated Metacore pathways are shown together with heatmap of reactome axon guidance genes. Fold change is shown in log2 scale (n=16 from 3 independent experiments, Benjamini Hochberg adjusted p-values, two-sided). C. Quantification of cell collisions in aligning fibroblasts (VCAF8 and VCAF2B) and non-aligning fibroblasts (CAF1, NF2.1 and CAF2) as either repulsion, alignment (collision guidance), adhere or ignore (n=124 collisions from three independent experiments). D. Analysis of CIL response in aligned (VCAF8, VCAF2B) and non-aligned (CAF1, NF2.1, CAF2) fibroblasts. Change in cell trajectory upon collision relative to the pre-collision path (blue arrow). (n=90 collisions from three independent experiments. p=5x10-5, one-sided t-test). E. Schematic of collisions between two cells. CIL is indicated by the repolarisation of the cell, relative to its pre-collision trajectory (angle between orange and orange-dashed line. Collision guidance is assessed by comparing the angle of approach (angle between orange and blue line) and departure, relative to its neighbour (angle between orange-dashed and blue line). Plot of example collisions. Diameter of dot indicates the strength of the repolarisation/CIL response. Collisions inside blue triangle reflect collision guidance events (departure angle is more than 10 degrees smaller than approach angle). F. Analysis of collision angles in aligned (VCAF8, VCAF2B) and non-aligned (CAF1, NF2.1, CAF2) fibroblasts. Collisions inside blue triangle reflect collision guidance events (n=124 collisions from two independent experiments). G. Quantification of collision guidance events (n=124 collisions from two independent experiments. p= 0.006, one-sided z-test). H. Model exploration of alignment as noise and collision guidance are co-varied. Text above indicates noise values fitted to experimental data for aligning (VCAF8, noise =0.1) and non-aligning (CAF1, noise=0.18) fibroblasts. Each square is the average of 10 independent simulations. I. Comparison of experimental alignment against model simulation with experimental persistence of VCAF8 (noise=0.1) and collision guidance=0.06 (indicated by black square in 3H). Experimental panels show bright field time-lapse microscopy of aligning VCAF8. Model panels show simulation of cell body organisation J. Model exploration of alignment as only CIL response and noise (persistence) are co-varied. Text above indicates noise values fitted to experimental data for aligning (VCAF8: noise =0.1) and non-aligning (CAF1: noise=0.18) fibroblasts. Each square is the average of 10 independent simulations.