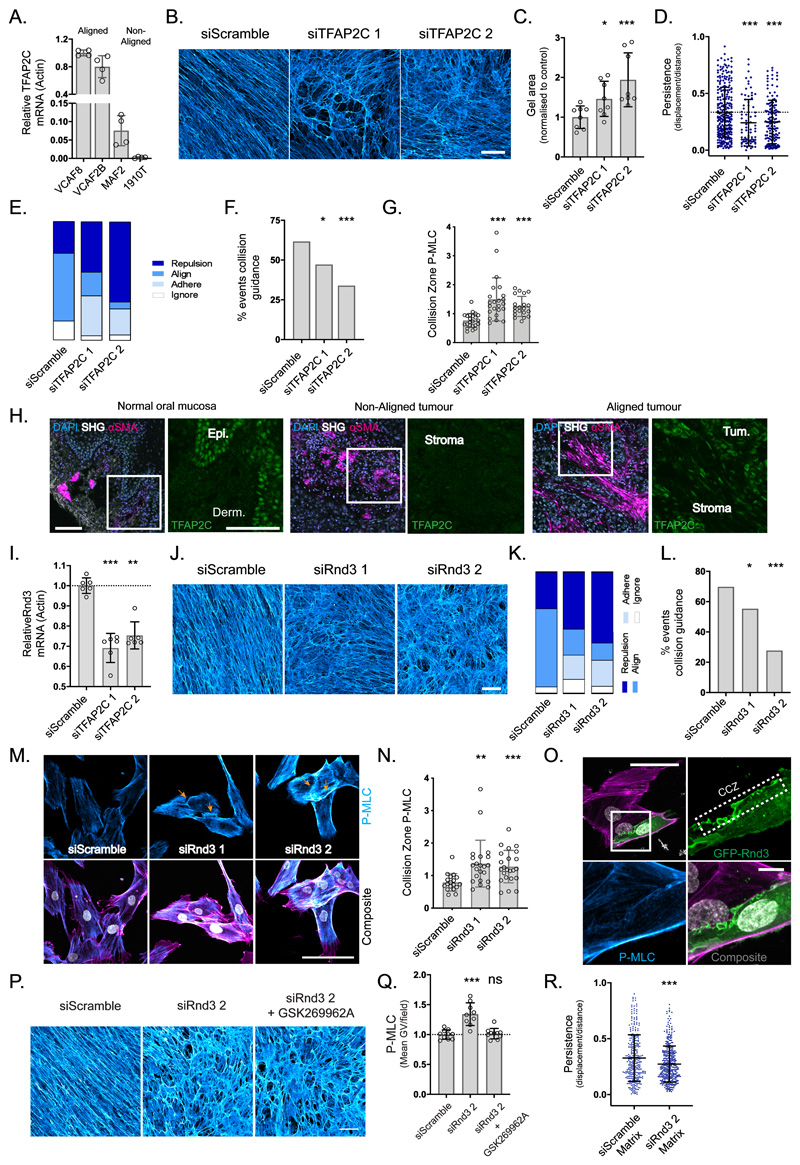
Figure 5. TFAP2C acts through RND3 to facilitate collision guidance and alignment.
A. qRT-PCR of TFAP2C gene expression in aligning vs non-aligning fibroblasts (n=16 from two independent experiments, mean and SD shown). B. FDMs from aligning VCAF8 transfected with two separate TFAP2C siRNA vs non-targeting control (scale bar:100μm, representative images of three fields of view from three independent experiments). C. Collagen gel contraction of aligning VCAF8 with TFAP2C knockdown vs control (n=24 from two independent experiments, p= 0.0265, p= 0.0028, unpaired two-tailed t-test, bars show mean and SD). D. Migratory persistence of aligning VCAF8 with TFAP2C knockdown vs control over 12 hr intervals (n=482 cells from two independent experiments, p=0.001, p=0.0001, unpaired two-tailed t-test, bars show mean and SD). E. Relative contribution of cell repulsion, alignment (collision guidance), adhesion or ignoring in aligning VCAF8 with TFAP2C depletion vs control (n=230 collisions in total from two independent experiments). F. Collision guidance events as a proportion of total collisions in TFAP2C knockdown vs control VCAF8 (n=230 collisions from two independent experiments. p=0.048, p= 0.00085, one-sided z-test). G. pS19-MLC at the cell-cell collision zone in TFAP2C knockdown vs control VCAF8. Values normalised to cell body pS19-MLC (n=66 cells, p<0.0001 unpaired two-tailed t-test, mean and SD are shown). H. Normal human oral mucosa and squamous cell carcinomas stained for DAPI (light blue), αSMA (magenta) and TFAP2C (green), with second harmonic imaging (grey), (scale bar: 100μm, from 6 independent samples). I. qRT-PCR analysis of relative RND3 gene expression in TFAP2C depleted cells vs control (n= 18 from three independent experiments, p<0.0001, p= 0.0014, unpaired two-tailed t-test, mean and SD are shown). J. Fibronectin stained matrices from VCAF8 with RND3 siRNA knockdown vs control (scale bar:100μm, representative images of three fields of view, from three independent experiments shown). K. Relative contribution of cell repulsion, alignment (collision guidance), adhesion or ignoring in VCAF8 with RND3 knockdown vs control (n=216 collisions in total from two independent experiments). L. Quantification of collision guidance events as a proportion of total collisions in RND3 knockdown vs control VCAF8 (n=216 collisions from two independent experiments, p=0.046, p<0.00001, one-sided z-test). M. F-actin (magenta), pS19-MLC (cyan) and DAPI (white) in RND3 depleted VCAF8 vs control (scale bar:100μm, representative images of three separate fields of view from two independent experiments shown). N. pS19-MLC values at the cell-cell collision zone in RND3 knockdown vs control VCAF8. Values normalised to cell body pS19-MLC (n=63 in total, p= 0.0027, p=0.0006, unpaired two-tailed t-test. Mean and SD are shown). O. VCAF8 transfected with eGFP-RND3 (green) and stained for pS19-MLC (cyan) and F-actin (magenta). Cell-cell collision zone (CCZ) (scale bar: 50μm, representative images from three independent experiments). P. Fibronectin stained FDMs from aligning VCAF8 with RND3 knockdown vs control, in the presence of absence of ROCK inhibitor (GSK269962A, 0.5nM) (scale bar=100μm, representative images of three fields of view, from two independent experiments shown). Q. pS19-MLC fluorescence intensity in VCAF8 from the above experiment (n=28 fields of view in total from two independent experiments, p<0.0001, p= 0.6866, bars show mean and SD). R. Migratory persistence of MDA-MB-231-GFP cells on FDMs derived from VCAF8 with Rnd3 knockdown vs control measured over 12 hr intervals (n=694 cells from 2 independent experiments, p=0.0001, two tailed unpaired t-test, bars show mean and SD).