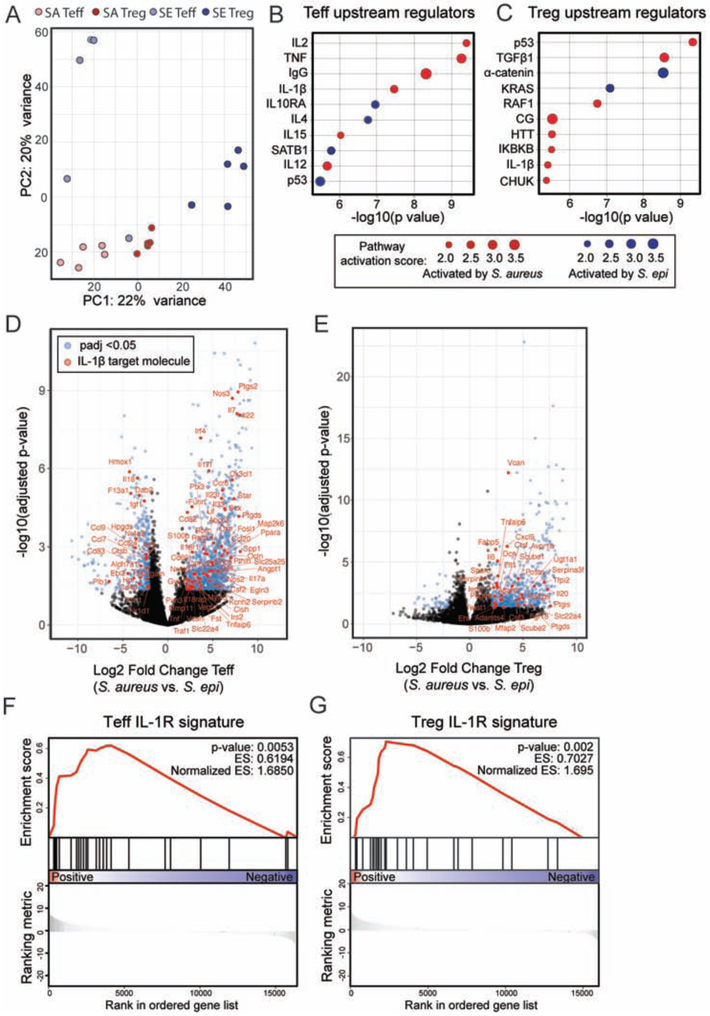
Figure 5: IL-1β is a key upstream regulator of the transcriptional signature of CD4+ T cells in the skin of neonates colonized with S. aureus as compared to S. epi.
WT pups were colonized with S. epi (SE) or S. aureus (SA) every two days from birth and skin Tregs (CD25hiICOShi) and CD4+ effector (CD25negICOSneg) T cells (Teff) were sorted from the skin at weaning age and processed for RNA sequencing. (A) Principal component analysis plot of the four sorted cell populations. (B) Upstream regulatory analysis was performed in Ingenuity on genes that were differentially expressed between SA vs. SE Teff and (C) SA vs. SE Tregs. Top ten regulators identified for each comparison for which there was a predicted direction of activation are shown in descending order of statistical significance. (D) Volcano plots showing padj value and fold change for genes differentially expressed by skin Teff and (E) Tregs from S. aureus vs. S. epi colonized mice. Blue dots represent genes with padj < 0.05 and > 2-fold difference in gene expression between groups. Genes in red are those annotated in Ingenuity as being IL-1β regulated. (F) Gene set enrichment analysis for the subset of these genes that are specifically regulated by IL-1β at the transcriptional level in S. aureus vs. S. epi Teff and (G) Tregs. See also Figure S5.