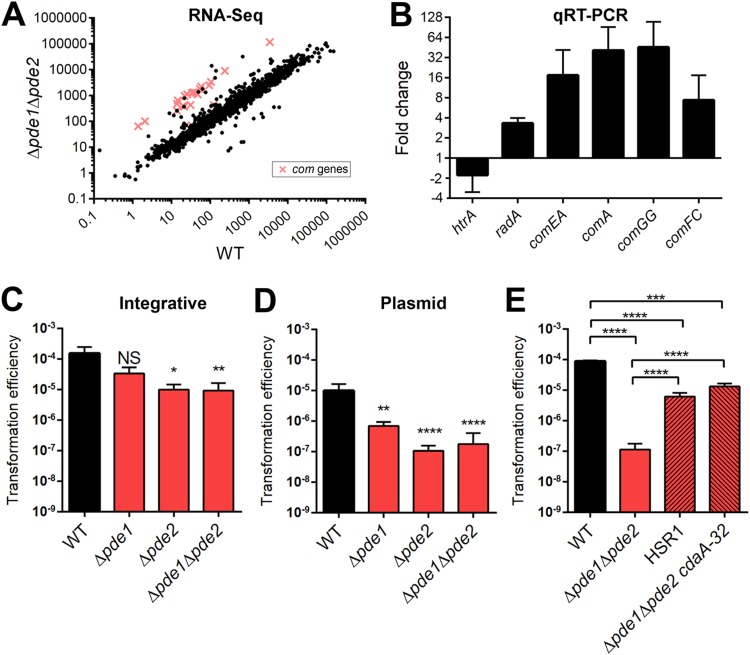
FIG 1.
Differential gene expression between the WT and Δpde1 Δpde2 strains determined using RNA-Seq and the effect of mutations in cdaA on transformation of the Δpde1 Δpde2 strain. (A) Scatterplot of RNA reads of the WT and Δpde1 Δpde2 strains determined using RNA-Seq. The com genes upregulated in the Δpde1 Δpde2 strain are indicated in red. Each spot or cross represents the average reads from three independent biological replicates. (B) Validation of RNA-Seq results by qRT-PCR. Fold changes of selected genes were determined from cDNAs generated from the same RNA samples as used in RNA-Seq. (C, D, and E) The WT, the pde mutants, HSR1, and the Δpde1 Δpde2 cdaA-32 strain were given CSP to induce transformation. Strains were transformed with either an integrative fragment with a Janus cassette (C) or plasmid pVA838 (D and E). Transformation efficiencies were calculated as output/input as described in Materials and Methods. Data shown are the means from three independent biological replicates. Error bars indicate the standard errors of the means (SEMs). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, not significant.