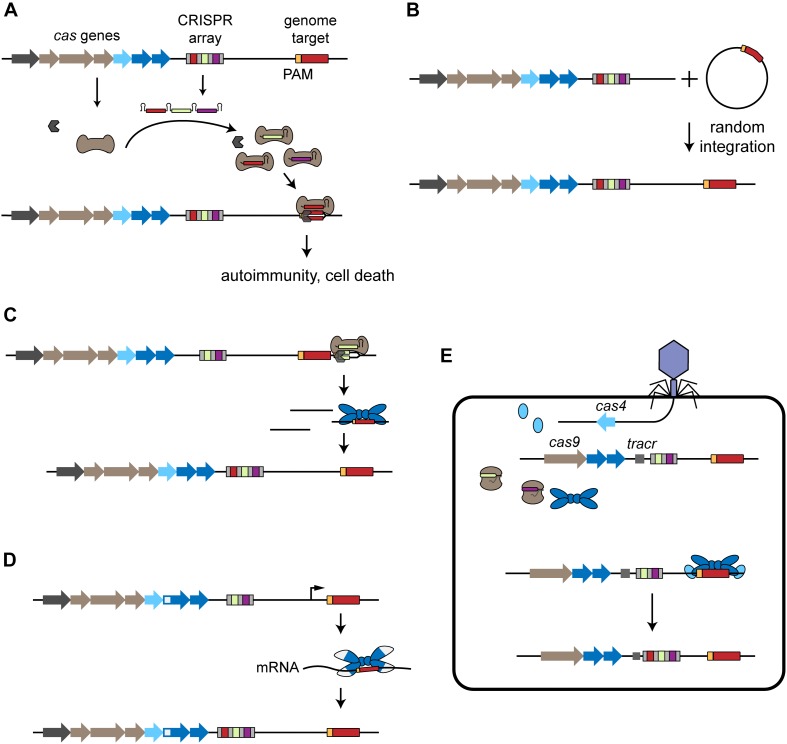
FIGURE 1.
Acquisition of self-targeting spacers. (A) Overview of self-targeting by CRISPR-Cas systems. The CRISPR array is transcribed and processed into individual crRNAs that form a ribonucleoprotein complex with the Cas effector proteins (brown). One of the crRNAs encodes a self-targeting spacer (red) that directs binding to the complementary protospacer sequence (red) flanked by a PAM (orange) located on the genome, leading to autoimmunity and cell death. (B) Mobile genetic elements harboring a CRISPR-Cas target sequence can be incorporated into the host chromosome, leading to self-targeting. (C) Primed acquisition. The CRISPR effector complex recognizes a target, potentially generating cleaved products. These products can then be incorporated into the CRISPR array by the acquisition complex (blue), leading to acquisition of self-targeting spacers. (D) Spacer acquisition from RNA. RT-Cas1 forms a complex with Cas2 (white and blue) and leads to incorporation of self-targeting spacers derived from the host’s RNA. (E) Virally driven acquisition of self-targeting spacers. The phage injects its genome into the host cell and the encoded cas4 is expressed. In cooperation with the host’s endogenous acquisition complex, the phage-derived Cas4 leads to the incorporation of genome-derived spacers into the host’s CRISPR array.