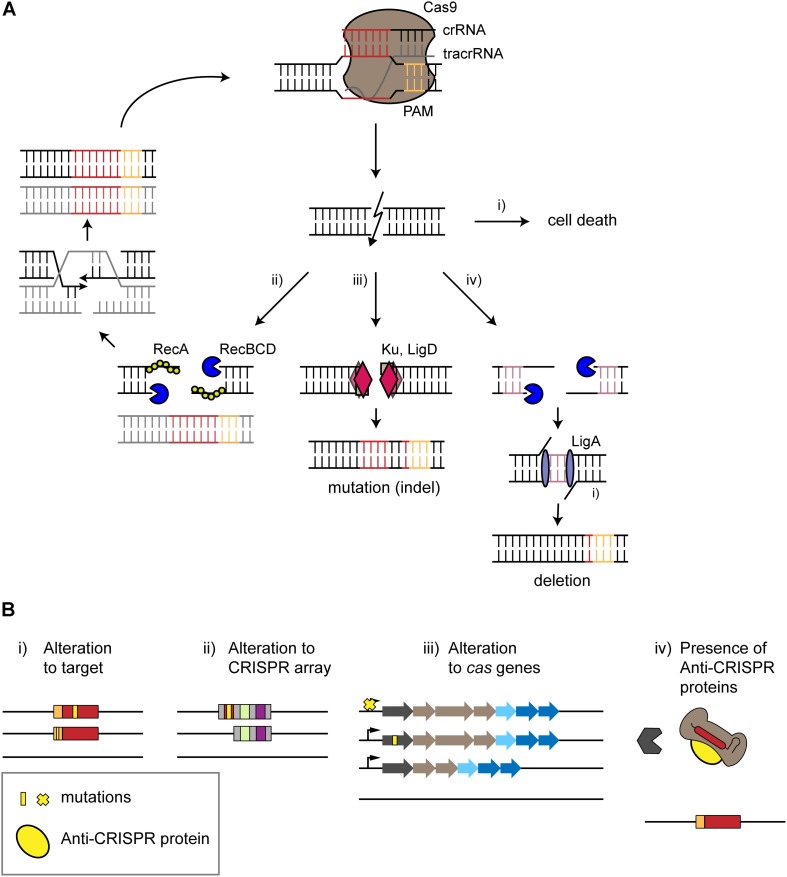
FIGURE 2.
Surviving self-targeting. (A) Survival of CRISPR-Cas targeting by intrinsic repair mechanisms. The type II effector protein Cas9 is used as an example. The CRISPR effector complex binds to its target in the genome (red) next to a PAM (orange), leading to a double-stranded break (DSB) causing different outcomes. (i) Cell death occurs if the break is not repaired. (ii) Homology-directed repair (HDR) restores the target site in the presence of an intact copy of the chromosome. DNA ends of the DSB undergo trimming in a Rec-dependent manner. HDR leads to the restoration of a chromosome that undergoes further attack by the CRISPR-Cas system. (iii) Non-homologous end joining leads to the formation of an insertion or deletion (indel). End joining is mediated by the repair proteins Ku and LigD. (iv) Alternative end-joining leads to deletions. DNA ends are trimmed by RecBCD until micro-homologous regions (purple) are reached. These regions are then ligated by LigA, resulting in deletions. (B) Escape from autoimmunity through mutations, deletions or active inhibition. (i,ii) mutations (yellow) or deletions within the protospacer, PAM or CRISPR array disrupts self-targeting. (iii) Mutation of the cas operon, inhibition of Cas expression or deletion of a cas gene or the entire locus can also prevent self-targeting. (iv) Anti-CRISPR proteins encoded within an integrated prophage can block CRISPR-Cas interference through different mechanisms, such as binding the Cas effector protein to prevent PAM recognition.