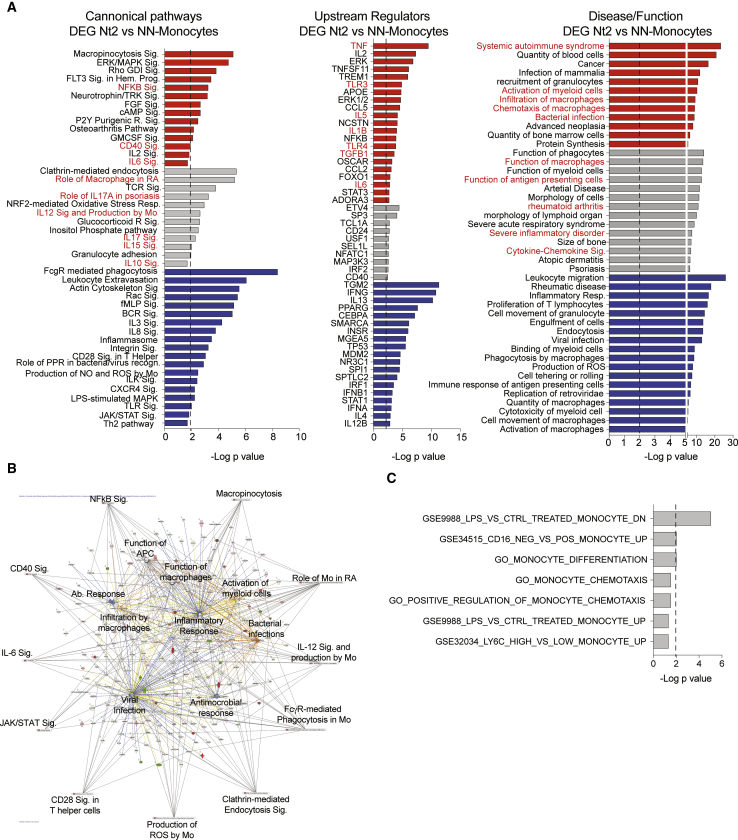
Figure 6.
Inflammatory Transcriptional Signatures in Mos from Nt2 Controllers
(A) Selected significant canonical pathways (left), upstream regulators (center), and diseases and functions (right) predicted by IPA from significant (FDR p < 0.05) DEGs in Mos from Nt2 versus NN controllers. Significance cutoff at −log p value = 2 was highlighted with a discontinuous line. Functions predicted to be upregulated, downregulated, or without predicted directional change are highlighted in red, blue, or gray, respectively.
(B) Network analysis of predicted canonical pathways (outer edges) and disease and functions (inside) for DEG in Mos from NT2 patients.
(C) Gene set enrichment analysis of significant DEGs from Nt2 versus NN controllers in n = 19 public gene sets related to Mo function and migration. Data correspond to −log FDR-corrected p values from selected predicted pathways.