Abstract
The Exo5 family consists of bi-directional, single-stranded DNA-specific exonucleases that contain an iron-sulfur cluster as a structural motif and have multiple roles in DNA metabolism. S. cerevisiae Exo5 is essential for mitochondrial genome maintenance, while the human ortholog is important for nuclear genome stability and DNA repair. Here, we identify the Exo5 ortholog in Schizosaccharomyes pombe (spExo5). The activity of spExo5 is highly similar to that of the human enzyme. When the single-stranded DNA is coated with single-stranded DNA binding protein RPA, spExo5 become a 5’-specific exonuclease. Exo5Δ mutants are sensitive to various DNA damaging agents, particularly interstrand crosslinking agents. An epistasis analysis places exo5+ in the Fanconi pathway for interstrand crosslink repair. Exo5+ is in a redundant pathway with rad2+, which encodes the flap endonuclease FEN1, for mitochondrial genome maintenance. Deletion of both genes lead to severe depletion of the mitochondrial genome, and defects in respiration, indicating that either spExo5 or spFENl is necessary for mitochondrial DNA metabolism.
1. Introduction
Exonuclease 5 (Exo5) was discovered in the 1980s in a biochemical screen for exonucleases in S. cerevisiae [1]. However, the catalytic activities and cellular functions of the enzyme were only recently analyzed, initially in S. cerevisiae, and subsequently in mammals [2, 3]. Both Exo5 orthologs are bi-directional, single-stranded DNA (ssDNA)-specific exonucleases. Human Exo5 (hExo5) slides along ssDNA prior to cleavage, generating products that are varying in size [3], whereas S. cerevisiae Exo5 (scExo5) preferentially cleaves dinucleotides as its major product [2].
A phylogenetic analysis of Exo5 shows a wide-spread occurrence of the enzyme, including fungi, plants and mammals. But not all species have an Exo5. This includes some well-studied model organisms. The enzyme is apparently missing from all insects and from nematodes, but not from the Priapulida worms. It is also missing from all birds, except for the small Palaeognathae superorder, which contains Emu and Ostrich (Supplementary Fig. S1). Therefore, it appears that Exo5 was lost on multiple occasions during evolution. In addition, there also is an apparent divergence in cellular function in different organisms. All members of the Saccharomycetales order of fungi show a strong mitochondrial localization sequence for Exo5, and indeed the S. cerevisiae EXO5 deletion is defective for mitochondrial DNA maintenance, but shows no apparent nuclear genome stability defect [2]. However, most orthologs outside the Saccharomycetales order lack a mitochondrial localization sequence, indicating that their main function may be nuclear (Supplementary Fig. S1). Accordingly, studies of the human Exo5 ortholog revealed it to be a protein with both cytoplasmic and nuclear localization, which is important for nuclear genome stability and DNA repair [3].
Mutations in the nuclease domain of human EXO5 are associated with testicular cancer and several other cancers [4]. However, such cases are infrequent in the cancer genome atlas (portal.gdc.cancer.gov). Much more common is the very high frequency of EXO5 silencing in many cancer types including glioblastoma (46%) and stomach adenocarcinomas (57%), such that EXO5 silencing makes up 95% of all EX05 alterations [5].
Given the importance of EXO5 as a putative tumor suppressor, we were interested in a genetically more tractable model system to investigate the in vivo functions of Exo5 outside of the Saccharomycetales order, in order to better understand the nuclear functions of Exo5. The fission yeast Schizosaccharomyces pombe is an attractive model organism, particularly since a large-scale proteomic study indicated that S. pombe Exo5 (spExo5) is localized to both the nucleus and mitochondrion [6].
In this paper we describe the biochemical and genetic characterization of spExo5. We observed that spExo5 is a bi-directional, ssDNA-specific exonuclease that shows a strikingly similar cleavage specificity to the previously characterized mammalian orthologs. Like with human Exo5, coating of the ssDNA with RPA restricts the activity of spExo5 to that with a unique 5’-3’ directionality. Fission yeast exo5Δ strains are sensitive to UV-irradiation, to alkylating agents, and in particular to interstrand crosslinking (ICL) agents. These results closely recapitulate our previous Exo5 knockdown experiment in human cells [3]. Finally, we demonstrate that spExo5 plays a redundant role with the flap endonuclease FEN1 (rad2+ in S. pombe), for the maintenance of mitochondrial DNA. Remarkably, an exo5Δ rad2Δ double deletion strain shows a severe decrease in mitochondrial DNA copy number and an associated defect in respiratory growth.
2. Materials and Methods
2.1. Materials and reagents
DNA modification and restriction enzymes were from New England Biolabs (Ipswich, MA). The drugs used were all from Sigma-Aldrich (St. Louis, MO): cis-platin (diamminecis-platinum(II) dichloride), cat. no. P4394; methyl methanesufonate (MMS), cat. no. 129925; 8-MOP (8-methoxypsoralen), cat. no. M3501. Monoclonal anti-humanPCNA antibody PC10 was purchased from Santa Cruz Biotechnology and used to probe for S. pombe PCNA as described [7]. Oligonucleotides were purchased from IDT (Coralville, IA) and purified by urea-polyacrylamide gel electrophoresis (PAGE): c81, TTGCCGATGAACTTTTTTTTTTGATCGAGACCTT; v81 AAGGTCTCCATCAAAAAAAAAAGTTCATCGGCAA. The polarity switch oligonucleotides were a gift from Tim Lohman of this Department. The 5’-32P-label was introduced on oligonucleotides c81 or 5’-5’ polarity switch oligonucleotide using [γ-32P] ATP and T4 Polynucleotide kinase. The 3’-32P-label was added to 3’-3’ polarity switch oligonucleotide by incubation with [α-32P] dATP with terminal deoxynucleotide transferase under manufacturers’ conditions.
2.2. S. pombe strains.
The exo5Δ strain was obtained from Bioneer Corporation. Double disruption mutants were either obtained by crossing the exo5Δ strain with strains containing other repair gene disruptions, or by direct integration of deletion marker cassettes. Genotypes of the double deletion strains were confirmed by diagnostic PCR. Strains are listed in Supplementary Table 1.
Growth of strains containing Exo+ plasmids was carried out on EMM media lacking Leu or Ura as appropriate, containing 5 μM thiamine (non-inducing conditions), or as otherwise indicated.
2.3. Plasmids
Plasmid pBL281 (S. pombe) contains the Schistosoma japonicum glutathione S-transferase (GST) gene fused to the N terminus of the Exo5 gene in vector pRS424-GALGST [8]. The GST tag is separated from the N terminus of the S. pombe, human, and mouse Exo5 gene by a recognition sequence for the human rhinoviral 3C protease (LEVLFQ/GP). Following cleavage by the protease, the N-terminal sequence of S. pombe Exo5 is extended with the GPEF sequence. Plasmid pBL281-207 has an active site mutation D207. Plasmid pBL281-176 has an active site mutation D176A. S. pombe expression plasmids pREP3x-spExo5 has spExo5 under the control of the repressible nmt (no message in thiamine) promoter. Plasmids and sequences are available upon request. The pBL288 plasmid series has the Exo5+ cDNA (or mutants) cloned into plasmid pREP3X (ars1 Len2+ nmt promoter and terminator) under control of the repressible nmt promoter. The pBL289 series has a triple FLAG tag fused to the C-terminus of the Exo5+ cDNA (or mutants) in the pBL288 plasmid backbone. The pBL298 series has GFP fused to the C-terminus of the Exo5+ cDNA (or mutants) in vector pREP4X (ars1 Ura4+ nmt promoter and terminator) under control of the repressible nmt promoter.
Oligonucleotides were purchased from IDT (Coralville, IA) and purified by urea-polyacrylamide gel electrophoresis (PAGE): c81, TTGCCGATGAACTTTTTTTTT TGATCGAGACCTT; v81 AAGGTCTCCATCAAAAAAAAAAGTTCATCGGCAA. The polarity switch oligonucleotides were a gift from Tim Lohman of this Department. The 5’-32P-label was introduced on oligonucleotides c81 or 5’-5’ polarity switch oligonucleotide using [γ−32P] ATP and T4 Polynucleotide kinase. While the 3’-32P-label was added to 3’-3’ polarity switch oligonucleotide by incubation with [α-32P] dATP with terminal deoxynucleotide transferase under manufacturers’ conditions.
2.4. Exo5 overproduction and purification.
Overproduction was carried out in S. cerevisiae strain FM113 (MATa ura-3-52 trp1-289 leu2-3112 prb1-1122 prc1-407 pep4-3) transformed with plasmid pBL281 (S. pombe Exo5), pBL281-207 (S. pombe Exo5-D207A), or pBL281-176 (S. pombe Exo5-D176). Growth, induction, and extraction were similar to the procedures described previously [9]. Cells were harvested and resuspended in 1/2 the volume of 3x buffer A (buffer A: 60 mM HEPES-NaOH [pH 7.8], 0.4 M sodium acetate, 0.1 mM EDTA, 0.01% polyoxyethylene (10) lauryl ether, 10 mM sodium bisulfite, 10 μM pepstatin A, 10 μM leupeptin), then frozen in liquid nitrogen. The frozen cell pellets were then blended in dry ice powder. All further preparation was carried out at 0-4 °C. After the thawing of the lysate, 10% glycerol, 1 mM dithiothreitol (DTT), 0.05 mM phenylmethylsufonyl fluoride (100 mM stock), and 150 mM ammonium sulfate (4 M stock), 0.45 % polymin P (10% stock, pH 7.3) were added to the lysate. The mixture was stirred for 15 minutes, the lysate was cleared at 40,000 × g for 30 minutes, and the supernatant was precipitated with 0.31 g/ml solid ammonium sulfate. The precipitate was collected at 40,000 × g for 30 minutes and then redissolved in buffer A0 (subscript indicates the sodium acetate concentration) 10% glycerol, and 1 mM dithiothreitol until the lysate conductivity was equal to that of buffer A400. The lysate was then used for batch binding to 1 ml of glutathione-Sepharose 4B beads (GE Healthcare), equilibrated with buffer A400, and gently rotated at 4 °C for two hours. GST-beads were collected at 1,000 rpm in a swinging-bucket rotor, followed by batch washes (3 × 20 ml of buffer A400). The beads were transferred to a 10 ml column and washed at 2.5 ml/min with 100 ml of buffer A400. The second washing was with 50 ml buffer A400 containing 5 mM Mg-acetate and 1 mM ATP. And the third washing used 50 ml of buffer A400 and 30 ml of buffer A200. Elution was carried out with a flow rate of 0.2 ml/min with buffer A200 containing 20 mM glutathione (pH adjusted to 8.0). The protein was eluted into four fractions. The fractions containing pure protein were incubated over night at 4 °C with 30U of rhinoviral 3C protease, diluted with A0 to equal A100 and loaded on a 1-ml Mono Q column. Protein was eluted with a linear gradient of buffer A100 to A1200. Pure Exo5 protein was eluted at 300mM to 400mM sodium acetate. Exo5 mutants were purified similarly throughout the procedures described.
2.5. SpExo5 Co-immunoprecipitation.
Co-IPs were carried out in S. pombe strain PYP102 (exo5Δ) by transformation of pBL289 or mutant. Cells were grown in Edinburgh minimal media (EMM) supplemented with 5 μM thiamine overnight. Cells were then harvested and resuspended in 1/2 the volume of 3x buffer A (buffer A: 60 mM HEPES-NaOH [pH 7.8], 150 mM sodium chloride, 0.1 mM EDTA, 0.05% polyoxyethylene (10) lauryl ether, 10 mM sodium bisulfite, 10 μM pepstatin A, 10 μM leupeptin), then frozen in liquid nitrogen. The frozen cell pellets were then blended in dry ice powder. All further preparation was carried out at 0-4 °C. After the thawing of the lysate, 10% glycerol, 1 mM dithiothreitol (DTT), and 0.5 mM phenylmethylsufonyl fluoride (100 mM stock) were added to the lysates. The lysates were cleared at 40,000 × g for 30 minutes, and the cleared lysate was then used for batch binding to FLAG-M2-beads (SIGMA), equilibrated with buffer A150, and gently rotated at 4 °C for two hours. FLAG-M2-beads were collected at 1,000 rpm in a swinging-bucket rotor, followed by batch washes (3 × 500 μl of buffer A150). The second washing was with 500 μl buffer A150 containing 5 mM Mg-acetate and 1 mM ATP. And the third washing used (3 × 500 μl of buffer A150). Elution was carried out in buffer A150 containing 150 μg/ml 3XFLAG peptide (SIGMA). The protein was eluted into two fractions and analyzed by SDS-PAGE followed by coomassie brilliant blue staining.
2.6. Exonuclease assays.
The standard 10 μl assay mixture contains 100 mM Tris-HCl (pH 7.8), 500 μg/ml bovine serum albumin, 5 mM DTT, 5 mM Mg-acetate, 50 mM NaCl, 50-100 fmol of 32P-end-labeled oligonucleotide substrate, and enzyme. Incubations were carried out at 30 °C for the indicated time periods. Deviations from the standard assay conditions are indicated in the legends of the figures. Reactions were stopped with 10 mM final concentration of EDTA in addition to 40% formamide and analyzed on a 17% PAGE-7 M urea electrophoresis. After the gels were dried, they were subjected to phosphorimager analysis.
2.7. S. pombe damage sensitivity assays.
Strains were inoculated in 5 ml of liquid yeast extract supplemented (YES) medium and grown overnight at 30 °C with shaking. The cells were then washed in phosphate-buffered saline and the cell density was determined by spectroscopy at 595nm. Serial 10-fold dilutions of late-log-phase cells, from 105 to 10 cells per spot, were spotted onto YES plates or YES plates containing the indicated concentrations of cis-platin (mM) or methyl methanesulfonate (MMS, %). Other YES plates were irradiated with the indicated dose of UV254 (J/m2) or ionizing radiation (Gy). For 8-methoxypsoralen treatment cells were grown to an OD of 0.4 and then treated with 5 μg/ml 8-methoxypsoralen for 15 minutes at 30 °C, followed by no irradiation (-), or irradiation at 365 nm with the indicated dose to activate the psoralen crosslinks. Cells were washed twice with PBS buffer and diluted for spotting on YES plates. Plates were grown for 2-3 days at 30°C and photographed.
3. Results and Discussion
3.1. Fission yeast and human Exo5 have comparable structures and activities
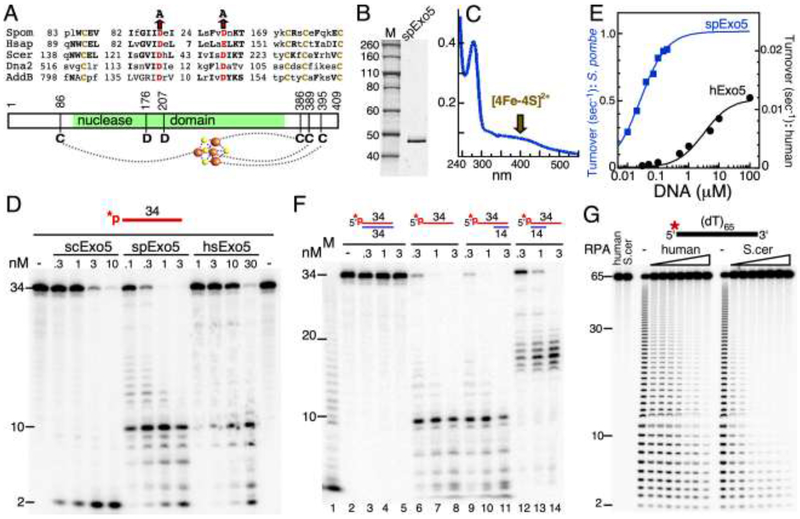
Budding yeast and human Exo5 show a conserved arrangement of cysteine residues that is also conserved in the nuclease domain of the B. subtilis AddAB recombinase and the Dna2 nuclease-helicase (Fig. 1A). These enzymes contain an iron-sulfur cluster moiety that is dependent on the presence of these cysteines [3, 10, 11]. Cys-Ser mutations in hExo5 resulted in a loss of iron-sulfur occupancy and an associated large decrease in enzymatic activity [3]. The conserved quartet of cysteine residues extends to fission yeast Exo5 (Fig. 1A). We overexpressed and affinity-purified spExo5 from S. cerevisiae (Materials and Methods). The purified enzyme shows an absorption at 410 nm, characteristic of a [4Fe-4S] iron-sulfur cluster (Fig. 1B,C).
Figure 1. Biochemical activities of spExo5.
(A) Conserved sequence motifs in the Exo5 family. Brown, cysteines that ligand the Fe-S cluster; red, catalytic residues. (B) 12% SDS-PAGE of purified spExo5. Staining was with Coomassie. (C) UV spectrum of purified spExo5; Absorption at 410 nm is indicative of the presence of a Fe-S cluster. (D) Comparison of activities of S. cerevisiae, S. pombe, and human Exo5 on a 34-nucleotide oligonucleotide. Standard assays contained 10 nM 5’-32P-labeled ssDNA substrate at the indicated concentrations of scExo5, spExo5, or hsExo5 for 4 min at 30 °C. Samples were analyzed on a 7 M urea-17% polyacrylamide gel. (E) Dependence of hExo5 and spExo5 activity on the concentration of 34-mer ssDNA. Data were fit to a Michaelis-Menten model. For spExo5, Km = 26 ± 2.2 nM and Vmax = 1 ± 0.2 sec−1; for hExo5, Km = 3,200 ± 800 nM and Vmax = 0.012 ± 0.002 sec−1. (F) Standard assays on 10 nM of the indicated fully dsDNA (lanes 2-5), ssDNA (lanes 6-8) and partial dsDNA (lanes 9-14) substrates, with indicated concentrations of spExo5. M, ladder from partial digestion of ssDNA with snake venom phosphodiesterase, a 3’-exonuclease. (G) 5’-directionality of spExo5 enforced by human or S. cerevisiae RPA. Standard assay mixtures used 10 nM 5’-32P-labeled (dT)65 substrate and 0.15 nM spExo5 with either no RPA, or 10, 12.5, 15, 20, 25, 30, or 40 nM of the indicated RPA for 5 min at 30 °C. Lanes 1 and 2 were control assays without Exo5 and with 40 nM of the indicated RPA.
We next investigated the enzymatic properties of spExo5 with the purpose of determining whether they were more like that of the S. cerevisiae or the human enzyme. The Exo5 family shows a strong conservation of active site residues (Fig. 1A). Mutation of either of two conserved aspartates that chelate the divalent metal required for catalysis resulted in a complete abrogation of enzymatic activity in vitro and a null phenotype in vivo for the budding yeast and the human enzymes [2, 3]. Consistent with these studies, mutation of either of the analogous active site aspartates to alanines (D176A, D207A), abrogated the nuclease activity of spExo5 (Supplementary Fig. S2A).
The Exo5 enzymes are designated as ssDNA-specific exonucleases due to their inability to degrade double-stranded DNA or circular ssDNA, i.e. they need an end as loading site [2, 3]. SpExo5 shares these properties with the budding yeast and mammalian family members. The enzyme is inactive on circular ssDNA (not shown) and on dsDNA (Fig. 1F). However, while budding yeast Exo5 generates predominantly dinucleotides as digestion products from a linear ssDNA substrate, both human and fission yeast Exo5 generate a large range of oligonucleotides (Fig. 1D). A comparison of the cleavage pattern generated by spExo5 with that by hExo5 revealed a remarkable similarity, with both enzymes exhibiting a dominant cleavage site that yielded a 10-mer. The two likely interpretations of that result are that either spExo5 prefers cutting at a distance ten nucleotides from the 5’-end, or that the ten-mer product results from preferential sequence or structure context. Cleavage of hompolymeric (dT)65 was entirely random (Fig. 1G, Supplementary Fig. S2B), which does not support the distance measuring model. Our interpretation of these data is that both forms of Exo5 load at one end and carry out a random walk along the ssDNA substrate prior to cutting, hence the name sliding exonuclease. However, despite the remarkable similarity in cleavage pattern (Fig. 1D), spExo5 is a much more active enzyme. At saturating DNA concentrations, the turnover number for spExo5 was about 100-fold higher than that of hExo5, 1 sec−1 vs 0.012 sec−1, respectively (Fig. 1E). An important caveat inherent to this comparison is that human Exo5 may require an as yet unidentified cofactor that stimulates its catalytic activity.
We next investigated the directionality of exonuclease action and the role of RPA in mediating directionality. The cleavage pattern observed in Fig. 1D could result from the enzyme loading at either the 5’- or 3’-terminus, followed by sliding along ssDNA prior to cleavage. The data in Fig. 1F suggest that spExo5 can load at either the 5’- or 3’-end. When the 3’-end was made inaccessible for Exo5 loading by hybridizing a complementary 14-mer (lanes 9-11), the cleavage pattern was similar to that of the ssDNA substrate. However, when the 5’-end was similarly made inaccessible (lanes 12-14), longer-sized products were generated, consistent with 3’-loading. Based on the disappearance of the 34-mer with increasing enzyme concentrations, the data also indicate that 5’-loading is more efficient than 3’-loading, by a factor of about five. An analysis of the activity of spExo5 on reverse polarity oligonucleotides is consistent with these conclusions (Supplementary Fig. S2B). The reverse polarity oligo(dT)70 substrates contain a polarity reversal in the middle, either a 3’-3’ or 5’-5’ intemucleotide linkage, such that the two ends of the oligo(dT) have either two 5’-ends or two 3’-ends, respectively. Similar to previously determined for the human enzyme [3], spExo5 showed activity on either substrate, with a preference for the 5’-ended substrate (Supplementary Fig. S2B).
In a physiologically relevant setting, ssDNA is normally coated with the ssDNA binding protein RPA. Both human Exo5 and the structurally related Dna2 nuclease assume a unique 5’-directionality when the ssDNA is coated with RPA [3, 12]. Likewise, coating of 5’-labeled (dT)65 with increasing concentrations of either human or S. cerevisiae RPA yielded progressively smaller oligonucleotides as products, which we propose to originate from 5’-loading and cutting near the 5’-end (Fig. 1G). Importantly, no products were seen in which the length of (dT)65 was reduced by just a few nucleotides, which would have been indicative of 3’-loading and cutting near the 3’-end. From these experiments we conclude that spExo5 shows remarkably similar enzymatic properties to hExo5 with regard to substrate cleavage specificities. The major difference is its increased catalytic efficiency. Therefore, we propose that S. pombe may be a tractable model system for understanding mammalian Exo5 function.
3.2. Mitochondrial and nuclear localization of spExo5
The genome stability and DNA damage phenotypes of a S. cerevisiae EXO5 deletion mutant are fully consistent with the sole mitochondrial localization of the protein [2]. On the other hand, depletion of human Exo5 is associated with nuclear chromosome integrity defects, and these are enhanced when cells are subjected to stress [3]. Because our genetic studies, to be described below, suggested the presence of both nuclear and mitochondrial defects in exo5 mutants, we have investigated the localization of Exo5 in more detail. A previous proteomic localization study already indicated that the protein was localized to both the nucleus and the mitochondrion [6]. However, a putative N-terminal localization signal for mitochondrial import is lacking (Supplementary Fig. S1; probability=0.00), using the Predotar prediction algorithm (https://urgi.versailles.inra.fr/Tools/Predotar). Nevertheless, mitochondrial localization might be mediated by another mechanism(s). S. pombe Exo5 contains several internal methionines in the unstructured ~80 amino acid long N-terminal region prior to the nuclease catalytic core. Alternative start site usage to direct subcellular localization is not an unusual mechanism. Human FEN1 has an alternative translation start site leading to a short, functional isoform that localizes to mitochondria, whereas the full-length form localizes to the nucleus [13]. Similarly, the nuclear and mitochondrial isoforms of the S. pombe Pif1 helicase result from alternative translation start sites [14]. Translation initiation at the Exo5-Met58 position was predicted by Predotar to have a mitochondrial localization probability of 0.18. Interestingly, Exo5 from two other Schizosaccharomyces species showed an even higher mitochondrial probability (0.6, 0.66) if initiation started at the analogous internal methionine. Therefore, we expressed in fission yeast three forms of Exo5: Exo5+, Exo5-M58A, and Exo5-Δ(1-57).
In order to carry out these experiments, Exo5+ was fused to a C-terminal 3xFLAG tag, and cloned into a S. pombe expression plasmid under control of the thiamine-inducible nmt promoter (see Materials and Methods). The nmt promoter shows a basal level of Exo5-FLAG expression in the presence of high levels of thiamine (5 μM)[15]. However, expression is strongly induced when thiamine is omitted from the media (Supplementary Fig. S3A). Under these highly inducing conditions, Exo5-FLAG levels were increased dramatically, and cells carrying the Exo5+ plasmid showed a negative growth phenotype (Supplementary Fig. S3B). The lethality phenotype resulting from Exo5 overexpression will be discussed in the next section. Addition to the growth media of 50 nM thiamine or higher was sufficient to relieve growth inhibition. At 50 nM thiamine, Exo5-FLAG levels are only slightly elevated from those at non-inducing conditions (Supplementary Fig. S3A). Therefore, all plasmid-based Exo5 studies (with or without C-terminal fusion tags or domains) were carried out with 5 μM thiamine in the media.
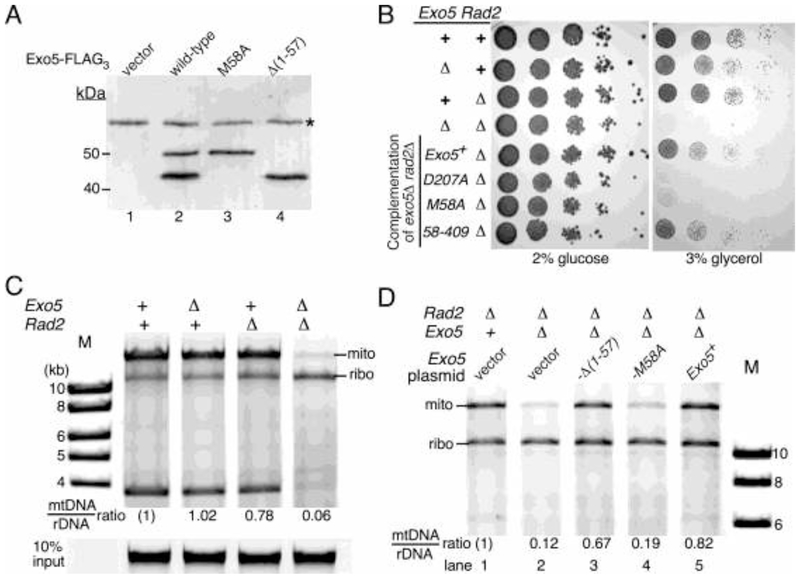
The affinity-purified wild-type Exo5-FLAG protein showed two prominent species by immunoblot blot analysis (Fig. 2A). The upper band is consistent with the predicted molecular weight of spExo5-3XFLAG protein (~50 kDa), while the lower band is consistent with that of a protein starting at Met58, followed by loss of a small signal peptide upon mitochondrial entry (~43 kDa). Importantly, the M58A mutant lacked the lower band, as one would expect if translation of the mitochondrial species started at Met58 with the M58A mutation eliminating this initiation. Conversely, the Δ(1-57) mutant showed only the lower band, further supporting our model.
Figure 2. Mitochondrial function of spExo5.
(A) Exo5Δ strains containing the pBL289 (exo5+-FLAG3) series of plasmids, placed under control of the nmt promoter, or empty vector were grown under non-inducing conditions (5 μM thiamine), and extracts subjected to Western analysis with 3xFLAG antibodies. *, non-specific band. (B) Isogenic strains with the indicated Rad2 or Exo5 deletions, and the Rad2Δ Exo5Δ double mutant containing the pBL288 series of plasmids (last 4 entries), were grown on selective media (-Leu), and ten-fold serial dilutions plated on rich media with either glucose or glycerol. Growth was for 3 days (YES) or for 6 days (YEG) at 30 °C. (C) Isogenic strains with the indicated Rad2 or Exo5 deletions were grown on rich media and chromosomal DNA isolated and digested with restriction enzymes AclI, AgeI, pstI, and PvuII, and separated on a 1% agarose gel. Staining was with GelRed and the fluorescence was recorded with a Typhoon phosphoimager in the fluorescence mode. The enzyme mixture cuts chromosomal DNA into small fragments but cuts the ribosomal DNA array only once per repeat (10.8 kb) and the mtDNA twice (15.7+3.8). The ratio of the 15.7/10.8 bands was quantified, with that of wild-type set to (1). Bottom, 10% of undigested input DNA. (D) Plasmid containing strains from (B) were grown on selective media with 5 μM thiamine. Chromosomal DNA was isolated and restriction enzyme digested, and separated on a 0.7 % agarose gel. Digestion and analysis was exactly as in (C).
We also expressed the same mutants with a C-terminal GFP tag under non-inducing conditions. Cells were fixed and observed by fluorescence microscopy. Wild-type Exo5+ showed diffuse cytoplasmic fluorescence and both nuclear and punctate mitochondrial fluorescence. The Exo5-M58A mutant showed diffuse cytoplasmic/nuclear fluorescence, but lacked punctate fluorescence suggesting its exclusion from the mitochondria. The Δ(1-57) mutant showed only punctate staining suggesting that this truncated form of Exo5 is solely localized to the mitochondria (Supplementary Table 2). Therefore, both sets of data are consistent with a model in which mitochondrial localization of spExo5 proceeds through translational initiation at Met58, whereas initiation at Met1 yields predominantly the cytoplasmic and nuclear forms.
3.3. Exo5 interacts with RPA and PCNA
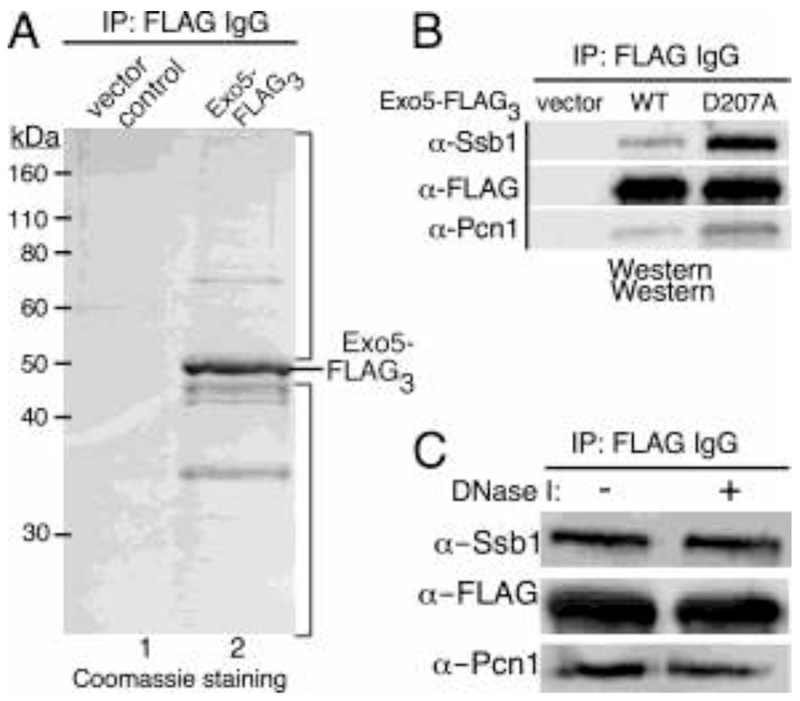
In order to potentially link spExo5 to specific DNA repair pathways, we performed co-immunoprecipitation experiments to find Exo5-interacting proteins (see Materials and Methods). Exo5-FLAG was expressed in a exo5Δ strain under non-inducing conditions. Cell extracts were subjected to co-immunopurification with 3xFLAG-beads and washed extensively prior to elution with a 3xFLAG peptide. The elutions were analyzed by SDS-PAGE followed by coomassie staining (Fig. 3A). While immunoprecipitation from the control extract showed very weak protein signals (lane 1), several proteins were easily detected after immunoprecipitation from the spExo5-3xFLAG-containing extract. The upper and lower sections of lane 2 (above and below the main spExo5 band) were cut out and analyzed by mass spectrometry. Mass finger printing identified several DNA replication and repair proteins with high significance. The full list of hits with high Mascot scores is in Supplementary Table 3. The top hits were two subunits of the single-stranded DNA binding protein RPA (Ssb1, Ssb2), two subunits of the 14-3-3 complex (Rad24, Rad25), and PCNA. Interestingly, the mitochondrial single-stranded DNA binding protein Rim1 was also identified as a significant hit, in accord with Exo5’s mitochondrial localization (Supplementary Table 3). To further test some of these interactions, we obtained antibodies against the S. pombe RPA70 subunit (Ssb1) and PCNA (Pcn1) [7, 16]. Indeed, both RPA and PCNA were detected by Western blot analysis in the Exo5-FLAG co-IPs, and not in the co-IPs from control extracts (Fig. 3B, Supplementary Fig. S3C). The interactions were not mediated through DNA as treatment with DNase I did not eliminate them (Fig. 3C, Supplementary Fig. S3D). Interestingly, co-IPs with the nuclease-defective form, exo5-D207A-FLAG, showed a strongly increased RPA and PCNA signal, suggesting that inactive Exo5-containg DNA repair complexes might be stalled on the DNA and therefore enriched during co-immunopurification (Fig. 3B). Previously, we showed biochemically that human Exo5 also interacts with human RPA [3]. The possible interaction of hExo5 with PCNA has not been investigated and neither form of Exo5 shows a PCNA consensus interaction (PIP) motif. The results from these experiments identify spExo5 through its interactions with both nuclear and mitochondrial DNA metabolic factors, but further work is required to characterize the nature and significance of these various interactions.
Figure 3. SpExo5 interacts with spRPA and spPCNA.
(A) Co-immunopurification with spExo5-FLAG3 from S. pombe cells grown under low-expressive conditions (5 μM thiamine). Elutions from the 3xFLAG beads were analyzed on a 10%-SDS PAGE gel and stained with coomassie brilliant blue. (B) Immunoblot analysis of co-immunopurifications of extracts from exo5Δ cells containing either empty vector, or plasmids containing 3xFLAG-Exo5 or the nuclease-defective 3xFLAG-spExo5-D207A mutant. Antibodies used against 3xFLAG (spExo5-FLAG3), spSsb1 (RPA70 subunit), and spPcn1 (PCNA). (C) Immunoblot analysis of extracts as in (B) either mock-treated or treated with DNaseI. The panels in (B) and (C) were assembled from the full Western blots shown in Supplementary Fig. S3C,D.
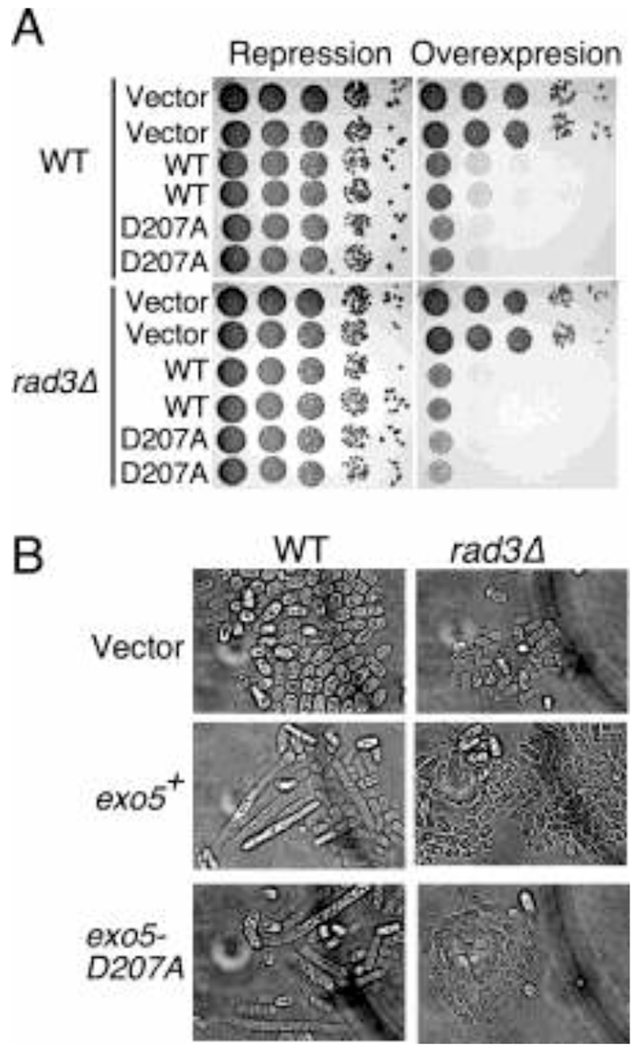
During our spExo5 expression experiments, we noticed that overexpression of Exo5+ from the strong nmt promoter in selective minimal media lacking thiamine caused lethality (Fig. 4A, Supplementary Fig. S3B). This lethality was not due to the nuclease activity of the protein, since overexpression of the nuclease-deficient mutant (exo5-D207A) showed similar lethality. Examination of the cell morphology revealed that the cells were elongated, indicative of checkpoint activation [17] (Fig. 4B). The DNA damage and replication checkpoints are mediated through activation of the Rad3 protein kinase, the ortholog of human ATR [17, 18]. However, while overexpression of exo5-D207A in a rad3Δ background eliminated the cell elongation phenotype, it did not suppress lethality (Fig. 4A,B). We hypothesize that checkpoint activation and lethality are the result of the increased Exo5 concentrations titrating away an essential replication factor(s), e.g. RPA or PCNA, from performing its proper functions during replication, but this hypothesis needs further testing.
Figure 4. Overexpression of SpExo5 causes checkpoint activation and cell death.
(A) Overexpression of spExo5 leads to cell death in S. pombe cells regardless of checkpoint proficiency. Cells either WT or rad3Δ containing pREP3x-spExo5 or pREP3x-spExo5-D207A were grown overnight in EMM-leu supplemented with 5μM thiamine. Cells were washed serially diluted on plates either containing 15 μM or 0 μM thiamine and incubated 2-3 day at 30 °C. (B) Light microscopy of cells from (A), either grown on plates containing 15 μM or 0 μM thiamine for 2-3 days.
3.4. S. pombe Exo5 and FEN1 play a redundant role in mitochondrial DNA maintenance
S. pombe is a petite negative yeast, which requires a functional mitochondrial genome for growth [19]. In glycerol-containing media, cells depend on respiration for energy source, which requires a fully functional mitochondrial machinery. Therefore, unlike in budding yeast, in fission yeast neither genotypic petite mutants (mutation or deletion of the mitochondrial genome) nor phenotypic petite mutants (failure to grow on a non-fermentable carbon source such as glycerol) are generally found. S. pombe exo5Δ strains are viable, indicating that spExo5 is not essential for mitochondrial genome stability (Fig. 2B). However, Exo5 is not the only 5’-nuclease present in mitochondria. The 5’-flap endonuclease FEN1 has a demonstrated involvement in mitochondrial genome maintenance in S. cerevisiae [20] and in human cells [21]. Therefore, it is reasonable to assume that S. pombe FEN1 (rad2+) fulfills a similar mitochondrial function. Interestingly, while neither the single exo5Δ nor rad2Δ mutant is associated with a detectable mitochondrial growth phenotype, the double mutant exo5Δ rad2Δ showed a failure to grow on media lacking a fermentable carbon source (Fig. 2B). The double mutant also showed a minor growth defect on rich media containing glucose (Fig. 5D). Exogenous expression of wild-type exo5+ restored the growth of these strains on glycerol plates and this restoration required the nuclease activity of Exo5 (Fig. 2B). Remarkably, expression of the short isoform exo5-(58-409), which shows mitochondrial localization, was able to restore growth of the exo5Δ rad2Δ mutant on glycerol plates, but expression of the exo5-M58A mutant, which eliminates translation of the short isoform, did not.
Figure 5. Exo5 genetic interactions in DNA repair pathways.
(A) Epistasis experiment with exo1Δ Ten-fold serial dilutions were plated on YES plates and exposed to UV-irradiation, or plated on YES supplemented with the indicated concentration of MMS. Right panel was rearranged for presentation; the original is shown in Supplementary Fig. S3E. (B) Sensitivity to acute psoralen treatment as described in the methods. Cells were washed twice with PBS buffer and diluted for spotting on YES plates. (C) Epistasis experiment with pso2Δ for sensitivity to chronic cis-platin treatment as described in the methods. Cells were spotted in ten-fold dilutions. (D) Epistasis experiments with rad2Δ for sensitivity to chronic cis-platin and MMS. (E) Epistasis experiments with fml1Δ for sensitivity to chronic cis-platin. Selected sections are shown; the full figure is in Supplementary Fig. S4B. (F) Epistasis experiments withpli1Δ for sensitivity to chronic cis-platin.
As an initial investigation into the mitochondrial defect of the exo5Δ rad2Δ double mutant, we isolated genomic DNA from an isogenic set of strains and digested the DNA with a set of restriction enzymes that allowed us to quantify the ratio of mitochondrial DNA to ribosomal repeat DNA (Fig. 2C). We observed a severe loss of mitochondrial DNA from the exo5Δ rad2Δ strain compared to the wild-type and single mutants, suggesting that Exo5 and FEN1 are redundantly required for mitochondrial DNA maintenance (Fig. 2C). In several experiments, the abundance of mitochondrial DNA in exo5Δ rad2Δ varied between 5-15% that of wild-type. No specific deletions were detected by altering the restriction enzymes used to digest the circular mitochondrial genome, nor were they detected by carrying out PCR with a series of primer sets that covered the circular genome (data not shown). Furthermore, the mitochondrial DNA loss phenotype was complemented by introduction of wild-type Exo5+ and of exo5-(58-409) on a plasmid, but not by the exo5-M58A mutant (Fig. 2D). These sets of data further support our model that the mitochondrial isoform of Exo5 results from initiation of translation at methionine-58. We conclude that those mitochondrial genomes that were defective due to the combined lack of FEN1 and Exo5 were lost from the population, perhaps by mitophagy [22]. The results from these experiments establish a redundant function in mitochondria for Exo5 and FEN1, presumably operating during the final steps of DNA replication in order to generate ligatable nicks.
3.5. Fission yeast Exo5 is important for DNA repair
In order to determine the possible participation of spExo5 in various nuclear DNA repair pathways, we carried out in vivo survival assays of exo5Δ mutant strains after exposure to DNA damaging agents. We detected no hypersensitivity of exo5Δ to γ-irradiation (Supplementary Fig. S4A), nor is a mre11Δ mutant, which is defective for X-ray repair, hypersensitized by the additional exo5Δ mutation (Supplementary Fig. 4B). However, the exo5Δ mutant is more sensitive than isogenic wild-type to UV-irradiation and alkylating agents (Fig. 5A). Furthermore, the deletion is particularly hypersensitive to interstrand crosslinking (ICL) agents such as 8-methoxypsoralen (Fig. 5B) and cis-platin (Fig. 5C). 8-methoxypsoralen intercalates into the DNA and forms interstrand crosslinks upon irradiation with visible light [23]. Remarkably, these results with DNA damaging agents recapitulate the knockdown experiments in human cells; hExo5-depleted cells showed hypersensitivity to UV, MMS, and cis-platin, but no hypersensitivity to γ-irradiation [3]. However, these phenotypes differ dramatically from those in S. cerevisiae, in which the exo5Δ mutant showed wild-type sensitivity to all DNA damaging agents tested, but was solely defective for mitochondrial DNA maintenance [2]. These damage-sensitivity phenotypes allowed us to use S. pombe as a tool to map genetic interactions between spExo5 and known components of various DNA repair pathways. We carried out DNA damage sensitivity experiments with a set of mutations in known DNA repair genes, with a particular focus on interstrand crosslink (ICL) repair pathways. The results of these epistasis experiments are shown in Figure 5 in and Supplementary Figures S4 and S5.
The strongest exo5Δ phenotype was observed when the deletion was combined with rad2Δ (FEN1). In the nucleus, FEN1 nuclease is required for Okazaki fragment maturation, ribonucleotide excision repair, and long patch base-excision repair [24] [25] [26]. The rad2Δ exo5Δ double mutant showed a synthetic growth defect even in absence of DNA damage (Fig. 5D). As described above, this growth defect could be the result of the loss of respiratory fitness of the double mutant, which is caused by severe loss of mitochondrial DNA (Fig. 2B,C). In addition, treatment of the rad2Δ exo5Δ strain with either MMS or cis-platin revealed strong hypersensitivity to these DNA damaging agents that is consistent with a synergistic interaction (Fig. 5D). Therefore, unless the loss of respiratory function sensitizes fission yeast to DNA damaging agents, these data indicate that Exo5 and FEN1 operate largely in different DNA repair pathways in the nucleus.
Fission yeast Exo1 exonuclease is involved in Okazaki fragment maturation, double-strand break repair, mismatch repair, and interstrand crosslink repair [27–29]. While the single exo1Δ and exo5Δ mutants showed a comparable sensitivity to UV, MMS and ICL agents, the double mutant exo1Δ exo5Δ showed an increased sensitivity to these agents, indicating that Exo1 and Exo5 repair these damages with partial redundancy (Fig. 5A, Supplementary Fig. S4E). However, the double mutant was not sensitive to γ-irradiation, again supporting a model in which Exo5 is not involved in double-strand break repair (Supplementary Fig. S4A). The UV sensitivity of a rad13Δ mutant, the 3’-endonuclease that functions in nucleotide excision repair (ortholog of human XPG) is increased in the double mutant with exo5Δ, suggesting that Exo5 does not have a function in nucleotide excision repair.
In S. pombe, there are three distinct, but partially redundant pathways for the repair of ICLs. One pathway has been established through the study of mutations in fan1+, fml1+, and rad51+, involving the Fanconi genes together with homologous recombination [30, 31]. A second pathway, acting in parallel with the Fanconi pathway, is defined by pso2+ and requires the nucleotide excision repair factor rad13+ (ortholog of human XPG) and homologous recombination (rad51+) [32, 33]. Thus, both the Fanconi and Pso2/nucleotide excision repair pathway also depend on homologous recombination. In addition, the SUMO E3 ligase pli1+ is involved in the resolution of DNA crosslinks in a separate pathway [31]. Given the sensitivity of exo5Δ for ICL agents, we carried out an epistasis analysis to determine in what ICL repair pathway Exo5 may function. The crosslink sensitivity of pso2Δ is substantially higher than that of exo5Δ (Fig. 5B, C), while the double mutant exo5Δ pso2Δ shows an increased sensitivity to cis-platin (Fig. 5C, Supplementary Fig. S5B). The Fanconi branch of ICL repair is represented by fml1+ and fan1+. Exo5+ is epistatic with fml1+, i.e. the double mutant is not more sensitive than the single mutants (Fig. 5E). Likewise, the exo5Δfan1Δ double mutant is not more sensitive than the single mutants (Supplementary Fig. S5A). These data suggest that Exo5 functions in the Fanconi pathway of ICL repair. In support of this conclusion are epistasis experiments with pli1+, which defines the third pathway through sumoylation [31]. Exo5Δ and pli1Δ show synergistic interactions indicating that they operate in different, competing pathways (Fig. 5F). Altogether, these data are consistent with a model in which Exo5 is required for ICL repair by the Fanconi pathway.
3.6. Conclusions
Our studies of Exo5 in three distantly related organisms, i.e. human and budding yeast, and here fission yeast, shows the remarkable versatility of this exonuclease. Exo5 has revealed an essential mitochondrial function, but no nuclear function in budding yeast, an important nuclear function and a redundantly essential mitochondrial function in fission yeast, and an important nuclear function in human. The fact that the Exo5 gene has been lost several times during evolution, e.g. in some superorders of worms and birds, but not in others, and in all insects, might suggest that its absence would be easy to compensate for by other exonucleases (Supplementary Fig. 1). Yet, the strong correlation of several types of cancers with EXO5 silencing suggests that at least in humans this is not the case [5].
S. pombe has provided a genetic tool for investigating the role of Exo5 in DNA repair. The DNA damage sensitivity of an exo5Δ strain closely recapitulates the hExo5 knockdown experiments in human cells[3]. Exo5Δ and rad2Δ (FEN1) show synergistic interactions in both nuclear and mitochondrial DNA metabolism (Fig. 2, 5D). FEN1 is required for Okazaki fragment maturation, long patch base-excision repair, and ribonucleotide excision repair [24, 26]. It is possible that, similarly to the exonuclease/helicase Dna2, Exo5 plays a redundant function in Okazaki fragment maturation when ssDNA flaps become bound by RPA and FEN1 is no longer able to act [34–37]. Exo5 and FEN1 also play an essential redundant function in the mitochondria in S. pombe. Both enzymes are single-strand specific exonucleases that may be required to remove single-strand DNA flaps during the termination of mitochondrial DNA replication. In addition, Exo1, another nuclease in the FEN1 family appears to perform complementary functions to Exo5 in the repair of UV damage and MMS damage (Fig. 5A).
Exo5Δ strains were particularly sensitive to interstrand cross-linking agents, such as cis-platin and 8-methyoxypsoralen. The repair of ICLs has been investigated in S. pombe [31]. These studies have started to delineate the pathways for repair of ICLs, but there are still many questions that need to be investigated. There are at least three pathways that have been identified in S. pombe for ICL repair. One of these pathways involves the nuclease Fan1 and the helicase Fml1, which are Fanconi anemia homologs; the second pathway is dependent on the nuclease Pso2 and the nucleotide excision repair endonuclease Rad13 (XPG); the third pathway requires the sumo ligase Pli1 [31]. Exo5 is epistatic with the proteins of the Fan1 dependent pathway of repair and is non-epistatic with the Pso2 nuclease and with Pli1 (Fig. 5C,E,F). Future studies will be required to determine the mechanistic function of Exo5 in ICL repair.
Supplementary Material
Highlights.
Exo5 is an understudied 5’-exonuclease that is downregulated in several cancers.
Fission yeast Exo5 is a good model system for the human enzyme.
Fission yeast Exo5 is functional in the nucleus and the mitochondrion.
Exo5 and FEN1 nuclease are redundantly required for mitochondrial DNA maintenance.
Exo5 mutants show defects in the Fanconi pathway of interstrand crosslink repair.
Acknowledgements
The authors thank Nick Rhind, Susan Forsburg, for providing plasmids, and Tim Lohman for a gift of reverse polarity oligonucleotides. The authors would also like to thank Nick Rhind, Susan Forsburg, Anthony Carr, Hiroshi Iwasaki, Lynda Groocock, Albert Pastink, Nancy Walworth, Daniel Teasley, Emily Higuchi, Michael Boddy, Randy Hyppa, Paul Russell, Toru Nakamura for providing S. pombe strains. The authors thank Hiroshi Iwasaki for providing Ssb1 antibodies used in this study. This work was supported in part by the US National Institutes of Health (GM118129 to P.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Burgers PMJ, Bauer GA, Tam L, Exonuclease V from Saccharomyces cerevisiae. A 5′-- -3′-deoxyribonuclease that produces dinucleotides in a sequential fashion, J. Biol. Chem, 263 (1988) 8099–8105. [PubMed] [Google Scholar]
- [2].Burgers PM, Stith CM, Yoder BL, Sparks JL, Yeast exonuclease 5 is essential for mitochondrial genome maintenance, Mol Cell Biol, 30 (2010) 1457–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sparks JL, Kumar R, Singh M, Wold MS, Pandita TK, Burgers PM, Human exonuclease 5 is a novel sliding exonuclease required for genome stability, J Biol Chem, 287 (2012) 42773–42783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Paumard-Hernandez B, Calvete O, Inglada Perez L, Tejero H, Al-Shahrour F, Pita G, Barroso A, Carlos Trivino J, Urioste M, Valverde C, Gonzalez Billalabeitia E, Quiroga V, Francisco Rodriguez Moreno J, Fernandez Aramburo A, Lopez C, Maroto P, Sastre J, Jose Juan Fita M, Duran I, Lorenzo-Lorenzo I, Iranzo P, Garcia Del Muro X, Ros S, Zambrana F, Maria Autran A, Benitez J, Whole exome sequencing identifies PLEC, EXO5 and DNAH7 as novel susceptibility genes in testicular cancer, Int J Cancer, 143 (2018) 1954–1962. [DOI] [PubMed] [Google Scholar]
- [5].Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, Fan H, Shen H, Way GP, Greene CS, Liu Y, Akbani R, Feng B, Donehower LA, Miller C, Shen Y, Karimi M, Chen H, Kim P, Jia P, Shinbrot E, Zhang S, Liu J, Hu H, Bailey MH, Yau C, Wolf D, Zhao Z, Weinstein JN, Li L, Ding L, Mills GB, Laird PW, Wheeler DA, Shmulevich I, Cancer N Genome Atlas Research, Monnat RJ Jr., Xiao Y, Wang C, Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas, Cell Rep, 23 (2018) 239–254 e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, Horinouchi S, Yoshida M, ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe, Nat Biotechnol, 24 (2006) 841–847. [DOI] [PubMed] [Google Scholar]
- [7].Reynolds N, Warbrick E, Fantes PA, MacNeill SA, Essential interaction between the fission yeast DNA polymerase delta subunit Cdc27 and Pcn1 (PCNA) mediated through a C-terminal p21(Cip1)-like PCNA binding motif, EMBO J., 19 (2000) 1108–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Walker J, Crowley P, Moreman AD, Barrett J, Biochemical properties of cloned glutathione S-transferases from Schistosoma mansoni and Schistosoma japonicum, Mol Biochem Parasitol, 61 (1993) 255–264. [DOI] [PubMed] [Google Scholar]
- [9].Bylund GO, Majka J, Burgers PM, Overproduction and purification of RFC-related clamp loaders and PCNA-related clamps from Saccharomyces cerevisiae, Methods Enzymol, 409 (2006) 1–11. [DOI] [PubMed] [Google Scholar]
- [10].Pokharel S, Campbell JL, Cross talk between the nuclease and helicase activities of Dna2: role of an essential iron-sulfur cluster domain, Nucleic Acids Res, 40 (2012) 7821–7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yeeles JT, Cammack R, Dillingham MS, An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases, J Biol Chem, 284 (2009) 7746–7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC, BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair, Genes Dev, 25 (2011) 350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kazak L, Reyes A, He J, Wood SR, Brea-Calvo G, Holen TT, Holt IJ, A cryptic targeting signal creates a mitochondrial FEN1 isoform with tailed R-Loop binding properties, PLoS One, 8 (2013) e62340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pinter SF, Aubert SD, Zakian VA, The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA, Mol Cell Biol, 28 (2008) 6594–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Forsburg SL, Comparison of Schizosaccharomyces pombe expression systems, Nucleic Acids Res, 21 (1993)2955–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Haruta N, Kurokawa Y, Murayama Y, Akamatsu Y, Unzai S, Tsutsui Y, Iwasaki H, The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro, Nat Struct Mol Biol, 13 (2006) 823–830. [DOI] [PubMed] [Google Scholar]
- [17].Jimenez G, Yucel J, Rowley R, Subramani S, The rad3+ gene of Schizosaccharomyces pombe is involved in multiple checkpoint functions and in DNA repair, Proc Natl Acad Sci U S A, 89 (1992) 4952–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Carr AM, Control of cell cycle arrest by the Mec1sc/Rad3sp DNA structure checkpoint pathway, Curr Opin Genet Dev, 7 (1997) 93–98. [DOI] [PubMed] [Google Scholar]
- [19].Haffter P, Fox TD, Nuclear mutations in the petite-negative yeast Schizosaccharomyces pombe allow growth of cells lacking mitochondrial DNA, Genetics, 131 (1992) 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kalifa L, Beutner G, Phadnis N, Sheu SS, Sia EA, Evidence for a role of FEN1 in maintaining mitochondrial DNA integrity, DNA Repair (Amst), 8 (2009) 1242–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM 3rd, Shen B, Demple B, Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria, Mol Cell Biol, 28 (2008) 4975–4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kanki T, Furukawa K, Yamashita S, Mitophagy in yeast: Molecular mechanisms and physiological role, Biochim Biophys Acta, 1853 (2015) 2756–2765. [DOI] [PubMed] [Google Scholar]
- [23].Cole RS, Psoralen monoadducts and interstrand cross-links in DNA, Biochim Biophys Acta, 254 (1971) 30–39. [DOI] [PubMed] [Google Scholar]
- [24].Liu Y, Kao HI, Bambara RA, Flap endonuclease 1: a central component of DNA metabolism, Annu Rev Biochem, 73 (2004) 589–615. [DOI] [PubMed] [Google Scholar]
- [25].Burgers PMJ, Kunkel TA, Eukaryotic DNA Replication Fork, Annu Rev Biochem, 86 (2017) 417–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, Crouch RJ, Burgers PM, RNase H2-initiated ribonucleotide excision repair, Mol Cell, 47 (2012) 980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tran PT, Erdeniz N, Symington LS, Liskay RM, EXO1-A multi-tasking eukaryotic nuclease, DNA Repair (Amst), 3 (2004) 1549–1559. [DOI] [PubMed] [Google Scholar]
- [28].Barber LJ, Ward TA, Hartley JA, McHugh PJ, DNA interstrand cross-link repair in the Saccharomyces cerevisiae cell cycle: overlapping roles for PSO2 (SNM1) with MutS factors and EXO1 during S phase, Mol Cell Biol, 25 (2005) 2297–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mimitou EP, Symington LS, DNA end resection: many nucleases make light work, DNA Repair (Amst), 8(2009)983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC, The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair, Mol Cell, 32 (2008) 118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fontebasso Y, Etheridge TJ, Oliver AW, Murray JM, Carr AM, The conserved Fanconi anemia nuclease Fan1 and the SUMO E3 ligase Pli1 act in two novel Pso2-independent pathways of DNA interstrand crosslink repair in yeast, DNA Repair (Amst), 12 (2013) 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Groocock LM, Prudden J, Perry JJ, Boddy MN, The RecQ4 orthologue Hrq1 is critical for DNA interstrand cross-link repair and genome stability in fission yeast, Mol Cell Biol, 32 (2012) 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lambert S, Mason SJ, Barber LJ, Hartley JA, Pearce JA, Carr AM, McHugh PJ, Schizosaccharomyces pombe checkpoint response to DNA interstrand cross-links, Mol Cell Biol, 23 (2003) 4728–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Murante RS, Rust L, Bambara RA, Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage, J. Biol. Chem, 270 (1995) 30377–30383. [DOI] [PubMed] [Google Scholar]
- [35].Bae SH, Bae KH, Kim JA, Seo YS, RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes, Nature., 412 (2001) 456–461. [DOI] [PubMed] [Google Scholar]
- [36].Ayyagari R, Gomes XV, Gordenin DA, Burgers PM, Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2, J Biol Chem., 278 (2003) 1618–1625. [DOI] [PubMed] [Google Scholar]
- [37].Kao HI, Veeraraghavan J, Polaczek P, Campbell JL, Bambara RA, On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing, J Biol Chem., 279 (2004) 15014–15024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.