Figure 2.
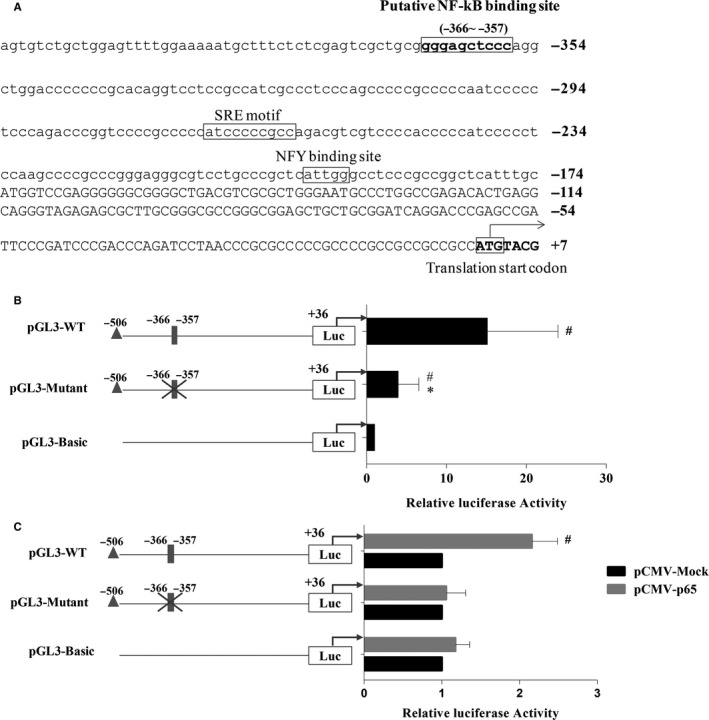
NF‐kB transactivated human PNPLA3 promoter through a putative NF‐kB binding site. A, Human PNPLA3 promoter upstream of the 5′UTR. The putative NF‐kB binding sites are highlighted with boxes; SREBP‐1c and NFY binding sites are underlined with a thick line. The ATG translation start codon where the A is numbered with 1 is indicated in bold and boxed. B, Relative luciferase activity of different PNPLA3 promoter‐reporter constructs. HepG2 cells were transfected with pGL3‐WT, PGL3‐Mutant and pGL3‐Basic reporter constructs for 24 h to measure the relative luciferase activities of PNPLA3 promoter by dual‐luciferase assays. Relative luciferase activity was corrected for Renilla luciferase activity and normalized to the pGL3‐Basic activity from three independent experiments. #P < .05 compared with pGL3‐Basic, *P < .05 compared with pGL3‐WT. C, NF‐kB‐driven relative luciferase activities from different PNPLA3 promoter‐reporter constructs in HepG2 cells. Each PNPLA3 promoter‐reporter construct was transiently cotransfected with pCMV‐Mock or pCMV‐p65 into HepG2 cells. Cell lysates were collected 24 h post‐cotransfection, and dual‐luciferase assays were performed. Relative luciferase activity was corrected for Renilla luciferase activity and normalized to the pCMV‐Mock activity from three independent experiments. # P < .05 compared with pCMV‐Mock. A grey‐filled rectangle represents the putative NF‐kB binding site, and a cross represents the mutation of the binding element
