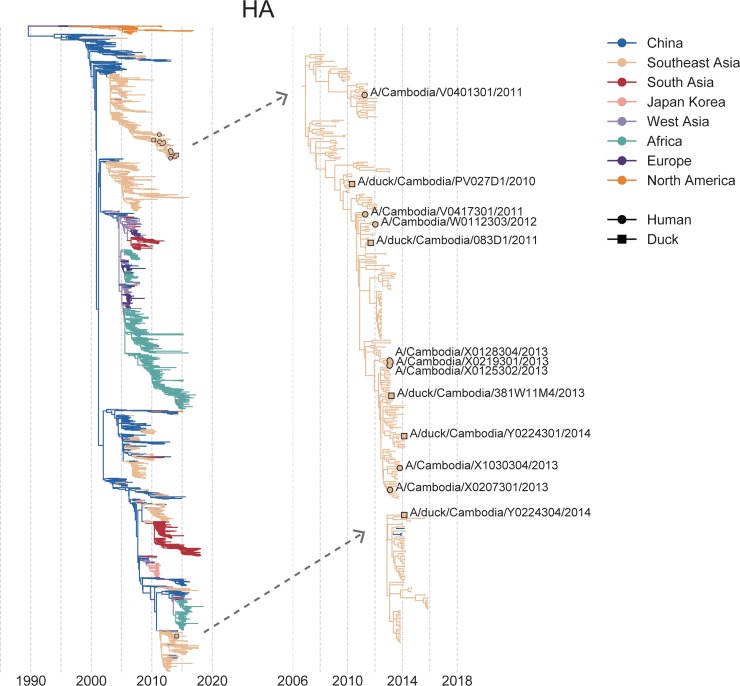
Fig 1. Phylogenetic placement of H5N1 samples from Cambodia.
All currently available H5N1 sequences were downloaded from the Influenza Research Database and the Global Initiative on Sharing All Influenza Data and used to generate full genome phylogenies using Nextstrain’s augur pipeline as shown in the trees on the left. Phylogenies for the full genome are shown in S2 Fig. Colors represent the geographic region in which the sample was collected (for tips) or the inferred geographic location (for internal nodes). The x-axis position indicates the date of sample collection (for tips) or the inferred time to the most recent common ancestor (for internal nodes). In the full phylogeny (left), H5N1 viruses from Cambodia selected for within-host analysis are indicated by tan circles with black outlines. The subtrees containing the Cambodian samples selected for within-host analysis are shown to the right and are indicated with grey, dashed arrows. In these trees, human tips are marked with a tan circle with a black outline, while duck tips are denoted with a tan square with a black outline. All samples from our within-host dataset are labelled in the subtrees with their strain name. Internal genes from samples collected prior to 2013 belong to clade 1.1.2, while internal genes from samples collected in 2013 or later belong to clade 2.3.2.1a. All HA and NA sequences in this dataset, besides A/duck/Cambodia/Y0224304/2014, belong to clade 1.1.2.