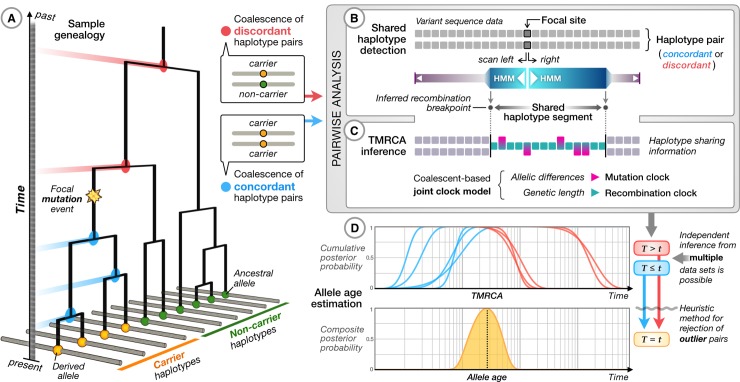
Fig 1. Overview of the GEVA method.
(A) At the chromosomal location of a variant, there exists an underlying (and unknown) genealogical tree describing the relationship between the samples. We assume that the derived allele (inferred by comparison to outgroup sequences) arose once in the tree. For concordant pairs of carrier chromosomes (yellow terminal nodes), their MRCAs (blue nodes) occur more recently than the focal mutation event. For discordant pairs of chromosomes, between the ancestral allele (green terminal nodes) and the derived allele, the MRCAs (red nodes) are older than the focal mutation. (B) For each pair of chromosomes (concordant and discordant), we use a simple HMM with an empirically calibrated error model to estimate the region over which the MRCA does not change; that is, the distance to the first detectable recombination event either side of the focal position along the sequence. From the inferred ancestral segment, we obtain the genetic distance and the number of mutations that have occurred on the branches leading from the MRCA to the sample chromosomes. (C) For each pair of chromosomes, we use probabilistic models (see S1 Text) to estimate the posterior distribution of the TMRCA, represented as cumulative distributions of having coalesced for concordant pairs (blue) and of having not coalesced for discordant pairs (red). (D) An estimate of the composite posterior distribution for the time of origin of the mutation is obtained by combining the cumulative distributions for concordant and discordant pairs. Informally, the mutation is expected to be older than concordant and younger than discordant pairs. In practice, this composite-likelihood–based approach results in approximate posteriors that are overconfident; hence, they are summarized by the mode of the distribution. Additional filtering steps are carried out to remove inconsistent pairs of samples (see S1 Text). GEVA, Genealogical Estimation of Variant Age; HMM, hidden Markov model; MRCA, most recent common ancestor; TMRCA, time to the most recent common ancestor.