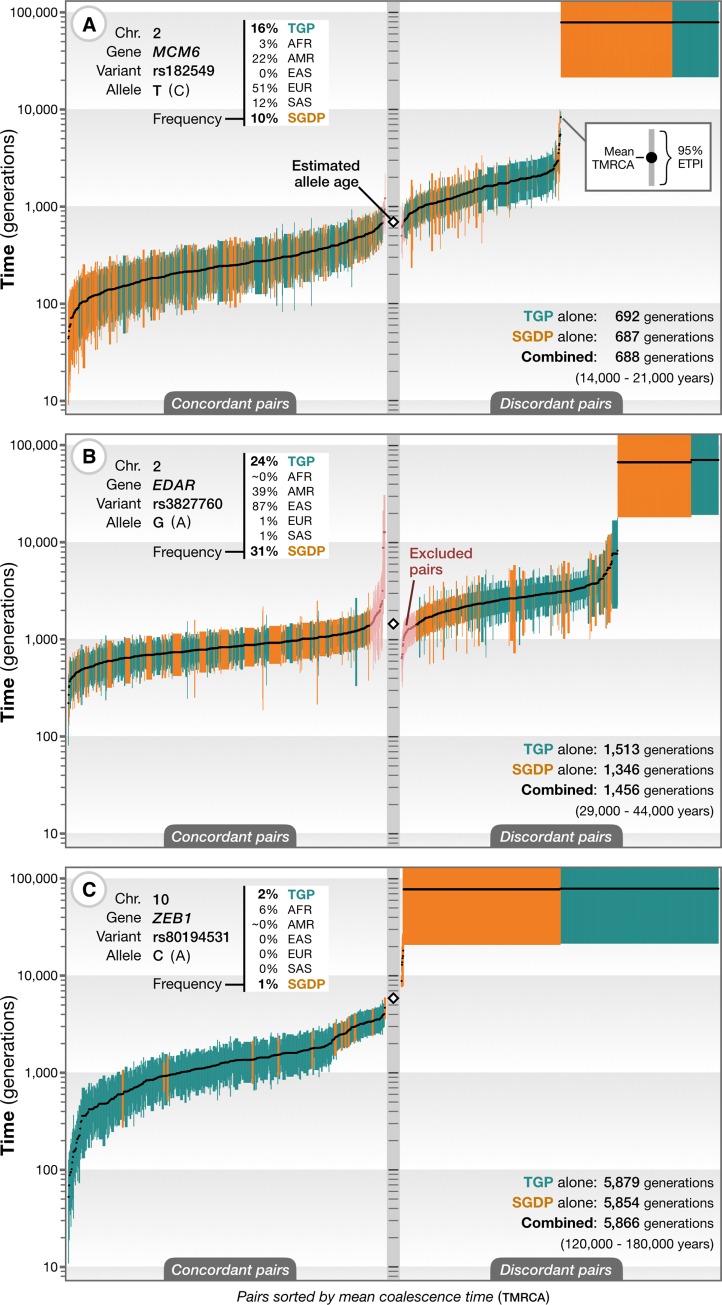
Fig 3. Application of GEVA to 3 variants of phenotypic and selective importance.
(A) Estimated TMRCAs for concordant (left) and discordant (right) pairs of chromosomes for the derived T allele at rs182549, which lies within an intron of MCM6 and affects regulation of LCT [33], which encodes lactase. Each bar reflects the approximate 95% credible interval (ETPI) for a pair, ordered by posterior mean (black dots). Data from the TGP (green) [2] and the SGDP (orange) [36] were used. The frequency of the variant in the SGDP, the TGP, and the different population groups in the TGP is shown (top left). The inferred allele age in generations from each data source and the combined estimate are shown (bottom right) and converted to an approximate age in years, assuming 20–30 years per generation. See https://human.genome.dating/snp/rs182549 for additional results. (B) As for panel A for the derived G allele of rs3827760, which encodes the Val370Ala variant in EDAR and is associated with sweat and facial and body morphology [41, 42]; also see https://human.genome.dating/snp/rs3827760. Our filtering approach is to remove the smallest number of concordant and discordant pairs necessary (shown in pink) to obtain concordant and discordant sets with nonoverlapping mean posterior TMRCAs. (C) As for panel A for the derived C allele of rs80194531, which encodes the Asn78Thr substitution in ZEB1, reported as pathogenic for corneal dystrophy [43]; also see https://human.genome.dating/snp/rs80194531. Abbreviations refer to ancestry groups. AFR, African; AMR, American; EAS, East Asian; EDAR, Ectodysplasin A Receptor gene; ETPI, equal-tailed probability interval; EUR, European; GEVA, Genealogical Estimation of Variant Age; LCT, Lactase gene; MCM6, Minichromosome Maintenance Complex Component 6 gene; SAS, South Asian; SGDP, Simons Genome Diversity Project; TGP, 1000 Genome Project; TMRCA, time to the most recent common ancestor; ZEB1, Zinc finger E-box–binding homeobox 1 gene.