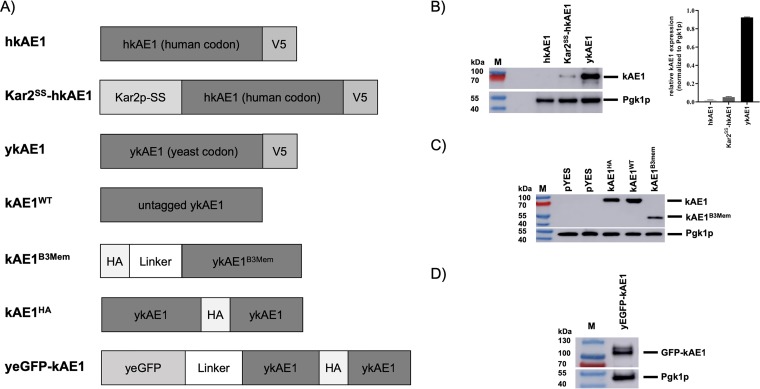
FIG 1.
Expression of various kAE1 variants in S. cerevisiae. (A) Schematic overview of full-length and truncated kAE1 fusion proteins constructed in this study. Positions of GGGGS linker (Linker), yeGFP, and epitope tags (V5 or HA) are indicated. In the truncated kAE1 version (kAE1B3Mem), the first 360 amino acids of the native AE1 sequence are deleted. (B) Expression profiles (left) and quantification (right, n = 2) of different V5-tagged kAE1 variants containing the original human (hkAE1) or yeast codon-optimized (ykAE1) DNA sequence. The ER signal sequence of Kar2p (Kar2SS) was added to the N terminus of the human kAE1 sequence to improve its ER targeting (Kar2SS-hkAE1). A protein standard (M) was used as a size determination marker, and phosphoglycerate kinase 1 (Pgk1p) served as a loading control. (C and D) Western blot analysis of yeast cells expressing yeast codon-optimized variants of untagged wild-type kAE1 (kAE1WT), kAE1 containing an HA tag within the third extracellular loop between Asn556 and Val557 (kAE1HA), truncated kAE1 (kAE1B3Mem) (C), or kAE1 with an N-terminal fusion to yeast enhanced GFP (yeGFP-kAE1) (D). Pgk1p served as a loading control.