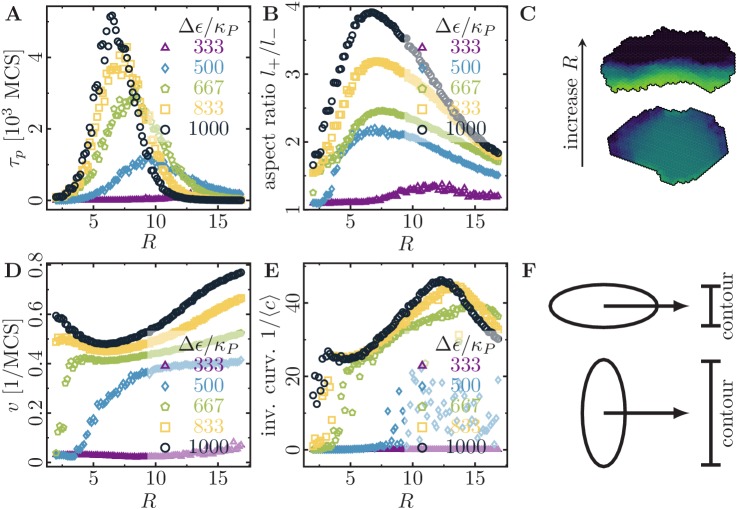
Figure 3. Migratory behavior of single cells as a function of the cell’s signaling radius at different values for the maximal cytoskeletal polarity .
(Stiffness parameters , ; average polarization field ; cell-substrate dissipation ; cell-substrate adhesion penalty ; cytoskeletal update rate ; independent simulations for each set of parameters). (A) The persistence times of directed migration of single cells exhibit a pronounced maximum at an optimal signaling radius, which depends on cell polarizability. (B) The shapes of single cells exhibit a pronounced maximal elongation at an optimal signaling radius, which depends on cell polarizability. (C) The signaling radius critically determines the synchronicity of internal cytoskeletal remodeling processes. Small signaling radii frequently lead to transient formation of mutually independent lamellipodia at different positions around the cell perimeter, thereby interrupting persistent motion (reducing persistence times of directed migration). Large signaling radii lead to structurally stable front-rear polarization profiles across the entire cell body (long persistence times of directed migration). Color code: cell polarization; cf. color bar in Figure 1C. (D) The speed of single cells does not drop to zero even when their persistence time of directed migration vanishes. This indicates single cell rotations. (E) The inverse curvature of the cell trajectories as a function of the signaling radius. (F) Depending on whether a cell migrates along its long axis (top) or short axis (bottom), it has to move a different projected contour length. If each protrusion takes roughly the same amount of time, then migration along the long axis (top; cell has to move a smaller projected contour length) allows for greater cell speeds than migration along the short axis (bottom; cell has to move a larger projected contour length).