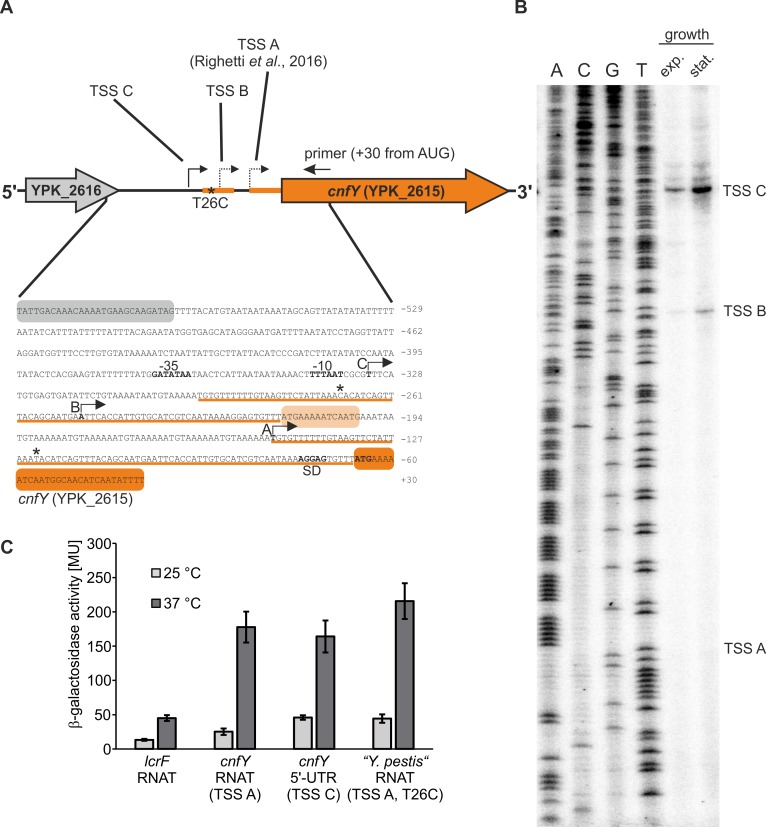
Fig 2. The cnfY RNAT functions regardless of the transcriptional site.
(A) Schematic representation of the cnfY (YPK_2615) gene locus. Transcriptional start sites (TSS) identified in a previous study [18] and in this study are marked by arrows. Additionally, the sequence of the intergenic region between YPK_2616 (coding for a putative transposase) and cnfY is displayed. The underlined sequences (orange line) represent the duplicated RNAT sequences. The sequence variation T26C observed in the duplicated sequences is marked by asterisks. (B) Identification of cnfY TSSs by primer extension. Total RNA was isolated from a Y. pseudotuberculosis YPIII culture grown at 37°C to exponential (OD600 = 0.5) and stationary growth phase (OD600 = 1.5). Primer extension was performed using a radiolabeled primer (binding is shown in A). The cDNA products were separated via polyacrylamide gel electrophoresis. The experiment was conducted in three biological replicates. (C) Y. pseudotuberculosis cnfY 5’-UTRs (+30 nt of the coding region; pBO3192, pBO6527) and the Y. pestis cnfY RNAT (-82 nt to +30 nt from AUG; pBO6523) were translationally fused to bgaB and tested for β-galactosidase activity. The lcrF RNAT was used as positive control (pBO3146). E. coli DH5α cells harboring the corresponding plasmid (technical triplicate per each construct) were grown to an OD600 = 0.5 at 25°C. Afterwards, transcription was induced with 0.01% (w/v) L-arabinose and the culture was split. One half remained at 25°C while the other was transferred into pre-warmed flasks (37°C). After 30 min of incubation, samples were taken for subsequent β-galactosidase assay. The displayed results represent the mean activities from three independent experiments (biological replicates). Mean standard deviations are indicated as error bars.