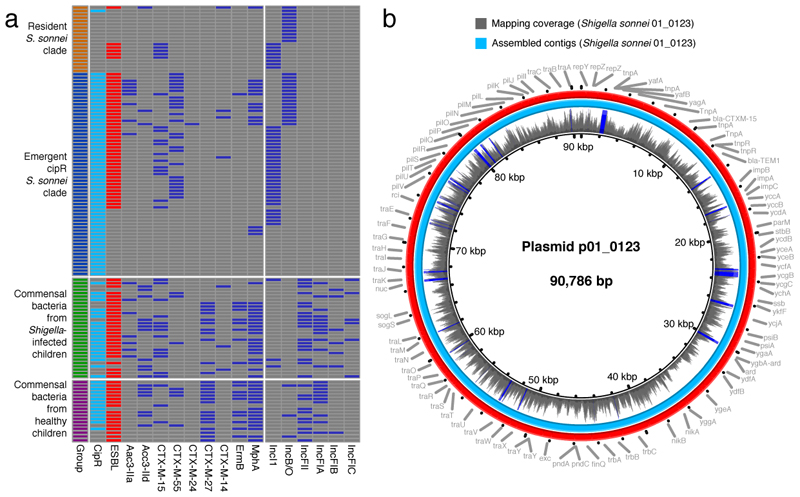
Figure 3. Antimicrobial resistance genes and plasmids in human commensal bacteria and Shigella sonnei.
a) The first column highlights the four different sample types. Fecal/rectal swab cultures with ciprofloxacin-resistant and ESBL-producing isolates are highlighted in turquoise and red (second and third columns, respectively). The remaining columns show the presence of key antimicrobial resistance genes and plasmid groups (blue) in commensal bacteria and S. sonnei. b) The central circle indicates the full sequence of plasmid p01-0123 assembled from Nanopore sequences of commensal E. coli. The next ring shows the depth of coverage from raw Illumina reads of ciprofloxacin-resistant Shigella sonnei strain 01-0123 mapped onto the central reference sequence. Graph height is proportional to the number of reads mapping at each nucleotide position in the reference genome from 0 to 30x coverage. Regions with plasmid coverage greater than 30x are shown as solid blue bands. The turquoise ring shows the BLASTN comparison between assembled sequences of Shigella sonnei strain 01-0123 and the central reference sequence. The red ring indicates the gene annotations of the central reference sequence.