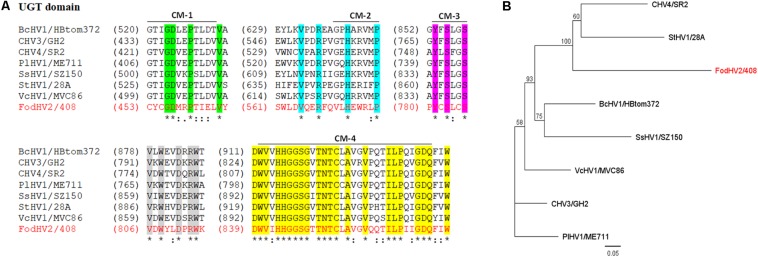
FIGURE 3.
Sequence comparison and phylogenetic tree of the UGT domain of Fusarium oxysporm f. sp. dianthi hypovirus 2 (FodHV2). (A) Multiple alignment of the aa sequence of the UGT domain between FodHV2/408 and selected viruses in the family Hypoviridae [BcHV1/HBtom372, CHV3/GH2, CHV4/SR2, PlHV1/ME711, SsHV1/SZ-150, StHV1/28A, and VcHV1/MVC86 (see Table 2 for complete name and accession number of the hypoviruses)]. Identical residues are indicated with asterisk and color shading; colons and dots indicate conserved and semi-conserved aa residues. CM: conserved motif sequences (1–4) of the UDP-glucosyltransferases superfamily. The alignements were performed using CLUSTALW with the program MAFFT v7. (B) Phylogenetic analysis based on the aa sequence of the UGT domain of FodHV2 and the mycoviruses used for the multiple alignment. The phylogenetic tree was constructed using the program Tree View of Geneious 8.1.5 package (Biomatters), and generated by the NJ method, with 1000 bootstrap replicates.