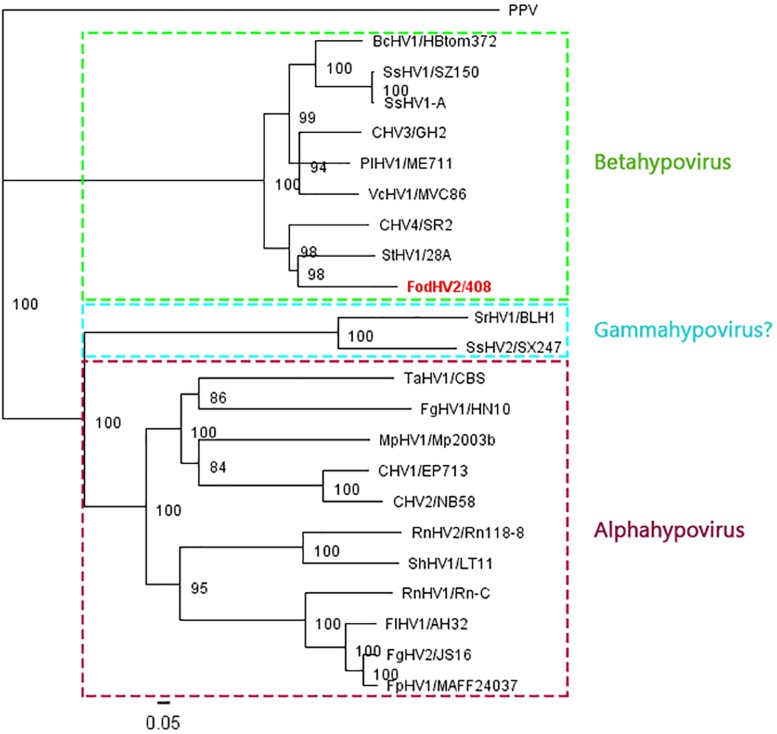
FIGURE 5.
Phylogenetic analysis of Fusarium oxysporm f. sp. dianthi hypovirus 2 (FodHV2) based on the aa sequence of the polyprotein. Neighbor-Joining consensus tree of FodHV2/408 and all the other hypoviruses [Botrytis cinerea hypovirus 1 (BcHV1/HBtom372; accession no. MG554632), Sclerotinia sclerotiorum hypovirus 1 (SsHV1/SZ-150; accession no. AEL99352), Sclerotinia sclerotiorum hypovirus 1-A (SsHV1-A; accession no. AWY10948), Chryphonectria hypovirus 3 (CHV3/GH2; accession no. AF188515), Phomopsis longicolla hypovirus 1 (PlHV1/ME711; accession no. KF537784), Valsa ceratosperma hypovirus 1 (VcHV1/MVC86; accession no. AB690372), Chryphonectria hypovirus 4 (CHV4/SR2; accession no. AY307099), Setosphaeria turcica hypovirus 1 (StHV1/28A; accession no. MK279474), Sclerotium rolfsii hypovirus (SrHV1/BLH1; accession no. AZA15168), Trichoderma asperellum hypovirus 1 (TaHV1/CBS; accession no. MK279475), Sclerotinia sclerotiorum hypovirus 2 (SsHV2/SX247; accession no. AIA61616), Fusarium graminearum hypovirus 1 (FgHV1/HN10; accession no. AZT88611), Macrophomina phaseolina hypovirus 1 (MpHV1/Mp2003b; accession no. ALD89099), Chryphonectria hypovirus 1 (CHV1/EP713; accession no. M57938), Chryphonectria hypovirus 2 (CHV2/NB58; accession no. L29010), Rosellinia necatrix hypovirus 1 (RnHV1/Rn-C; accession no. PRJNA485481), Rosellinia necatrix hypovirus 2 (RnHV1/Rn118-8; accession no. BBB86776), Sclerotinia homoeocarpa hypovirus 1 (ShHV1/LT11; accession no. MK279473), Fusarium langsethiae hypovirus 1 (FlHV1/AH32; accession no. YP_009330037), Fusarium graminearum hypovirus 2 (FgHV2/JS16; accession no. PRJNA485481), Fusarium poae hypovirus 1 (FpHV1/MAFF24037; accession no. BAV56305), and the plant potyvirus Plum pox virus (PPV) as an out-group] based on the full length aa sequence of the viral polyprotein. The color boxes indicate the different proposed genera of “Alphahypovirus,” “Betahypovirus,” and “Gammahypovirus.” The phylogenetic tree was constructed using Tree View of Geneious 8.1.5 package (Biomatters), and generated by the NJ method with 1000 bootstrap replicates.