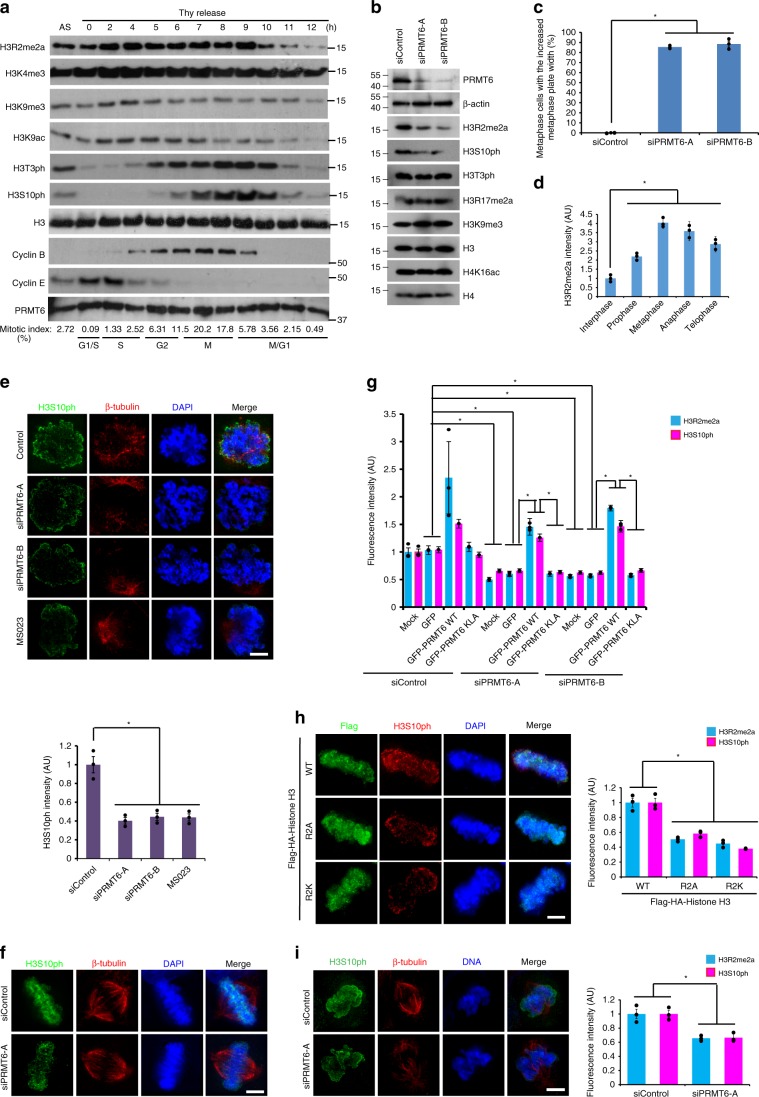
Fig. 1. PRMT6 facilitates H3S10 phosphorylation via H3R2 dimethylation.
a Levels of the indicated proteins from synchronous HeLa S3 cells were analyzed by fluorescence-activated cell sorting (FACS) and Western blotting. b Seventy-two hours after transfection with PRMT6-specific siRNA, HeLa cells were harvested and the levels of the indicated proteins were analyzed. c Seventy-two hours after transfection with PRMT6-specific siRNA, the metaphase cells, in which the metaphase plate width was increased in PRMT6-depleted HeLa cells, were quantified and plotted. (n = 150 metaphase cells from three independent experiments). d The intensity of H3R2me2a was quantified in fixed HeLa cells and plotted (n = 30 cells per each phase from three independent experiments). e, f HeLa cells were treated with siPRMT6 or MS023 as a PRMT6 inhibitor and stained with anti-H3S10ph antibody in early prometaphase and metaphase. The intensity of H3S10ph in prophase cells was quantified and plotted (n = 30 cells from three independent experiments). g The intensity of H3R2me2a and H3S10ph in GFP-positive cells was quantified and plotted (n = 30 cells from three independent experiments). h Twenty-eight hours after Flag-histone H3 WT or mutant (R2A or R2K) transfection, the H3S10ph and H3R2me2a intensities in Flag-positive cells were quantified and plotted (n = 30 cells from three independent experiments). i The intensity of H3R2me2a and H3S10ph in PRMT6-depleted RPE1 cells was quantified and plotted (n = 30 cells from three independent experiments). Scale bars, 5 μm. Error bars, SEM. P values were calculated by two-tailed Student’s t-tests (c, d, e, h, and i; *p < 0.01) or two-way ANOVA (g; *p < 0.01). Source data are provided as a Source Data file.