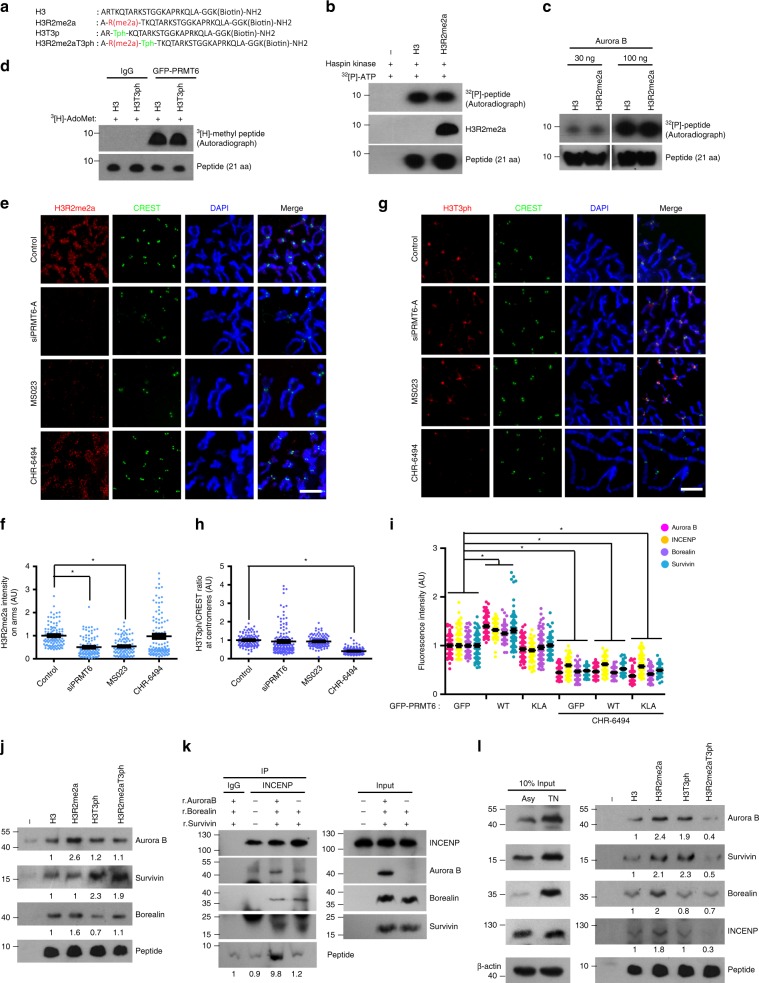
Fig. 5. Aurora B preferentially binds to H3R2me2a and recruits CPC components.
a The amino acid sequences of modified histone H3 peptides. b In vitro Haspin kinase assay with the 21-aa peptide. c In vitro Aurora B kinase assay with the 21-aa peptide for phosphorylation. d In vitro PRMT6 methyltransferase assay. e–h The cells were treated with the siPRMT6, the PRMT6 inhibitor MS023, or the Haspin kinase inhibitor CHR-6494. The intensities of H3R2me2a (f) and H3T3ph (h) in the chromosome arm and centromere were analyzed by immunofluorescence microscopy of nocodazole-arrested HeLa chromosome spreads and were plotted (n = 100 chromosomes from three independent experiments). i After transfection of the indicated plasmids, the HeLa cells were treated with a Haspin kinase inhibitor, CHR-6494, for 5 h and the intensity of CPC components was analyzed for 100 centromeres from three independent experiments. j Recombinant CPC proteins were incubated with biotinylated histone peptides and pulled down with streptavidin-agarose beads, and the binding was visualized by immunoblotting. k Lysates of TN-arrested Aurora B-depleted cells were supplemented with recombinant Survivin and Borealin proteins with or without recombinant Aurora B protein. The lysates were incubated with antibodies against INCENP and the H3R2me2a peptide, and pulldown was conducted with agarose A beads. The binding was visualized by immunoblotting. The asterisk denotes the light chain of antibodies. l Lysates of TN-arrested mitotic HeLa cells were incubated with biotinylated histone peptides and pulled down with streptavidin-agarose beads, and the binding was visualized by immunoblotting. Relative band intensities of band were measured with image processing software (Image Studio ver5.0). Error bars, SEMs. Scale bars, 5 μm. Source data are provided as a Source Data file. (Student’s t-test *p < 0.01).