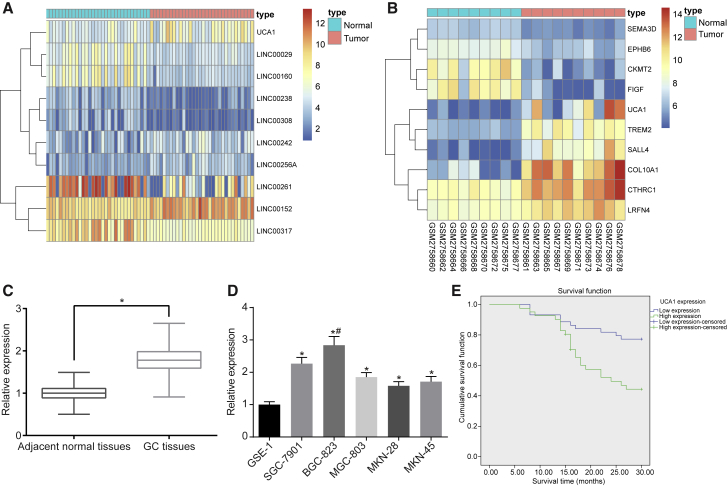
Figure 1.
UCA1 Was Highly Expressed in GC Tissues and GC Cells (MKN-28, MKN-45, SGC-7901, BGC-823, and MGC-803)
(A) Heatmap of GSE65801. *p < 0.05, versus the adjacent normal tissues. (B) Heatmap of GSE103236. The abscissa represents the sample number, the ordinate represents the differentially expressed gene name, the upper right histogram is the color order, the top-down color change represents the expression value of the chip data from large to small, each rectangle corresponds to the expression value of a sample, and each column represents the expression level of all genes in each sample. The left tree chart shows the clustering analysis results of different genes from different samples. The top bar indicates the type of sample, the upper right box indicates the color reference of the sample, the blue is the normal control sample, and the red is the tumor sample. (C) UCA1 was highly expressed in GC tissues (n = 85). (D) UCA1 was highly expressed in GC cells (MKN-28, MKN-45, SGC-7901, BGC-823, and MGC-803). *p < 0.05, versus the GSE-1 cell line; #p < 0.01, versus the GSE-1 cell line. (E) Kaplan-Meier analysis shows that expression of UCA1 is negatively correlated with GC patient survival (the measurement data in GC cell lines were analyzed by one-way ANOVA, and the measurement data in GC and adjacent normal tissues were analyzed by paired t test, each experiment was conducted three times independently).