Figure 6.
Predictive Signatures and AML Biology
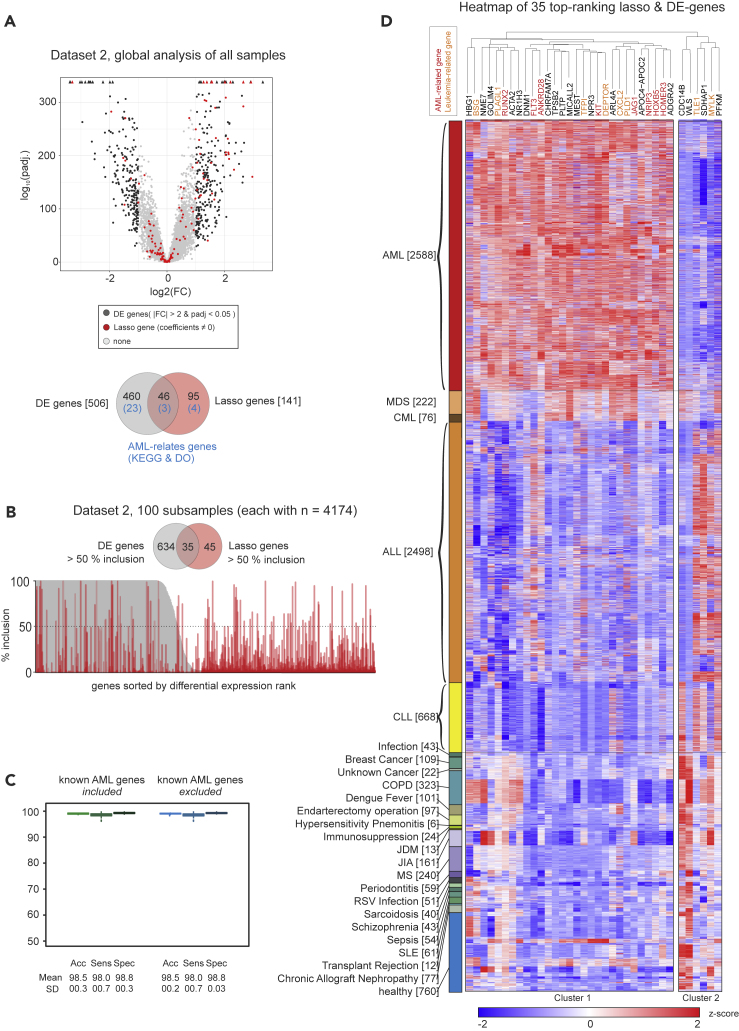
(A) Volcano plot of global differentially expressed (DE) genes and genes of the lasso model (“lasso genes”) in dataset 2, and Venn diagram indicating the overlap of both gene sets and the genes included in the KEGG pathway or the disease ontology term “AML.”
(B) Inclusion plot of DE genes and lasso genes in 100 random permutations of dataset 2. The plot is sorted according to DE gene rank, and a Venn diagram shows the overlap between genes with a minimum of 50% inclusion.
(C) Boxplot of accuracy, sensitivity, and specificity of the predictive model trained and tested on random subsets of dataset 2 with inclusion of all genes of the dataset and without 155 genes known to be relevant for AML biology. Error bars depict the standard deviation.
(D) Heatmap and hierarchical clustering of z-scaled expression values of 35 genes with >50% inclusion both in lasso and DE genes, as shown in (B). Genes with known associations with AML are marked red; genes associated with other types of leukemia are labeled in orange.
See also Figure S15.