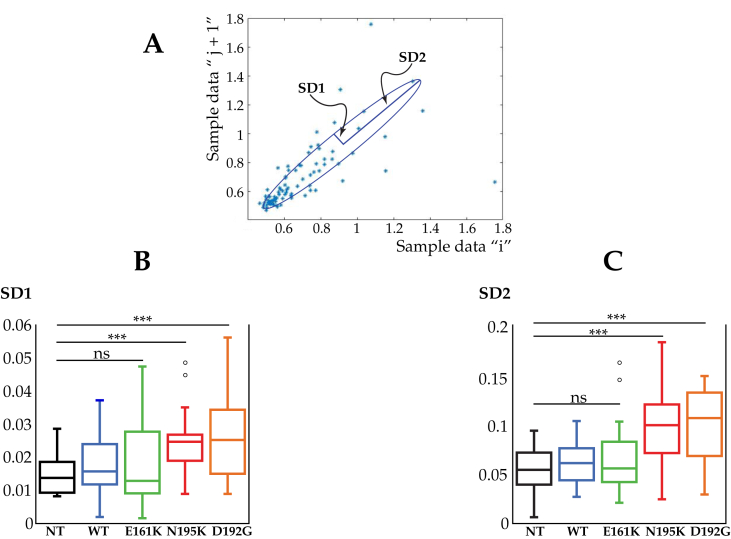
Figure 2.
Beating rate variability by Poincaré plot of LMNA mutant cardiomyocytes. (A) A typical Poincaré Plot of RR intervals for NT cardiomyocyte. The peak interval (RR n) is displayed on the x-axis and the extent of the following interval (RR n+1) on the y-axis. All points described by successive beats of equal duration (RR n = RR n+1) are situated on the identity line. The points below this line represent all shortenings of the interval between 2 consecutive beats (RR n > RR n+1) while the points above the identity line correspond to all prolongations (RR n < RR n+1). (B) Analysis of the beating rate variability of LMNA mutant cardiomyocytes. SD1 (the standard deviation of the instantaneous - short term - beat-to-beat variability and (C) SD2 (the standard deviation of the long-term interval variability) for NT, WT and LMNAE161K, LMNAN195K, and LMNAD192G mutations. ***Represents p < 0.0001, (t-test).