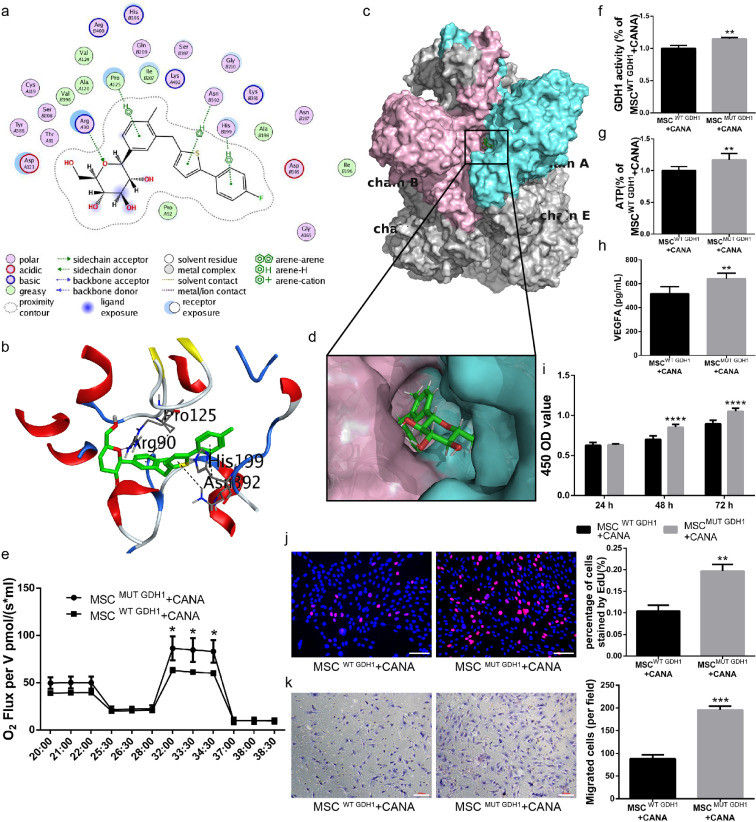
Fig. 8.
CANA interacts with the GDH1 hexamer by binding to a pocket formed by Chains A and B. (a) Docking simulations between CANA and GDH1 were performed using MOE software, which provided a 2D diagram of CANA/GDH1. The chemical structure of CANA is shown in the center, and its key interacting amino acids are shown around the structure. The annotations for the atom properties and interaction patterns were shown below. A and B in the circle of residue names denote Chains A and B of the GHD1 hexamer. The proximity contour indicates the amount of space that is available for substitutions on the ligand. Ligand exposure: the smudges drawn behind some of the ligand atoms indicate the solvent-accessible surface area of the ligand; receptor exposure: the size and intensity of the blue circle behind the residues correlated with the extent of solvent exposure of the corresponding residues as a result of the present of the ligand. (b) A 3D model of the docking poses of CANA with the GDH1 hexamer is shown. The purple dotted lines indicate hydrogen bonds; the green dotted lines indicate arene interactions; and the distance (expressed in Å) between atoms is provided for each hydrogen bond and arene interaction. Oxygen atoms are marked in red; nitrogen atoms are marked in blue; and sulfur atoms are marked in yellow. (c) CANA docks into the binging pocket of the GDH1 hexamer formed by Chains A (whose surface is shown in aquamarine) and B (whose surface is shown in pink); the surface of the other chains of the GHD1 hexamer are shown in gray. (d) A magnified view of the docking interface is displayed on the lower panel. (e) MSCs were first transfected with shRNA targeting mouse GDH1. These cells were then infected with recombinant lentiviruses carrying wild-type (WT) or H199A and N392A mutant (MUT) GDH1 cDNA (MSCsWT GDH1 or MSCsMUT GDH1). The resulting MSCsWT GDH1 and MSCsMUT GDH1 were incubated with CANA (10 μM, 48 h), and their OCRs were assayed under both basal and maximal conditions (i.e., under CCCP treatment; n = 3). (f) MSCsWT GDH1 and MSCsMUT GDH1 were incubated with CANA (10 μM, 48 h), their GDH1 activity was evaluated using a spectrophotometric assay (n = 3), and (g) their intracellular ATP levels were determined via luciferin/luciferase-based assays. (h) The cells were treated as indicated in (f), and 5 × 106 cells were cultured in CANA-free DMEM supplemented with 2% FBS for an additional 24 h. The culture supernatants were collected, and the concentration of VEGFA in the supernatant was analysed by ELISA. (i) The cells were treated as indicated in (f), and the proliferation of MSCs was evaluated by a CCK-8 assay (n = 3) and (j) an EdU staining assay (Scale bar: 100 μm). (k) The migration ability of the cells was evaluated using a Transwell migration assay (Scale bar: 100 μm). Data are means ± SDs, *P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001.