Figure 5 |. Molecular structures of bacterial components in situ determined by three-dimensional averaging of individual volumes extracted from whole cell tomograms.
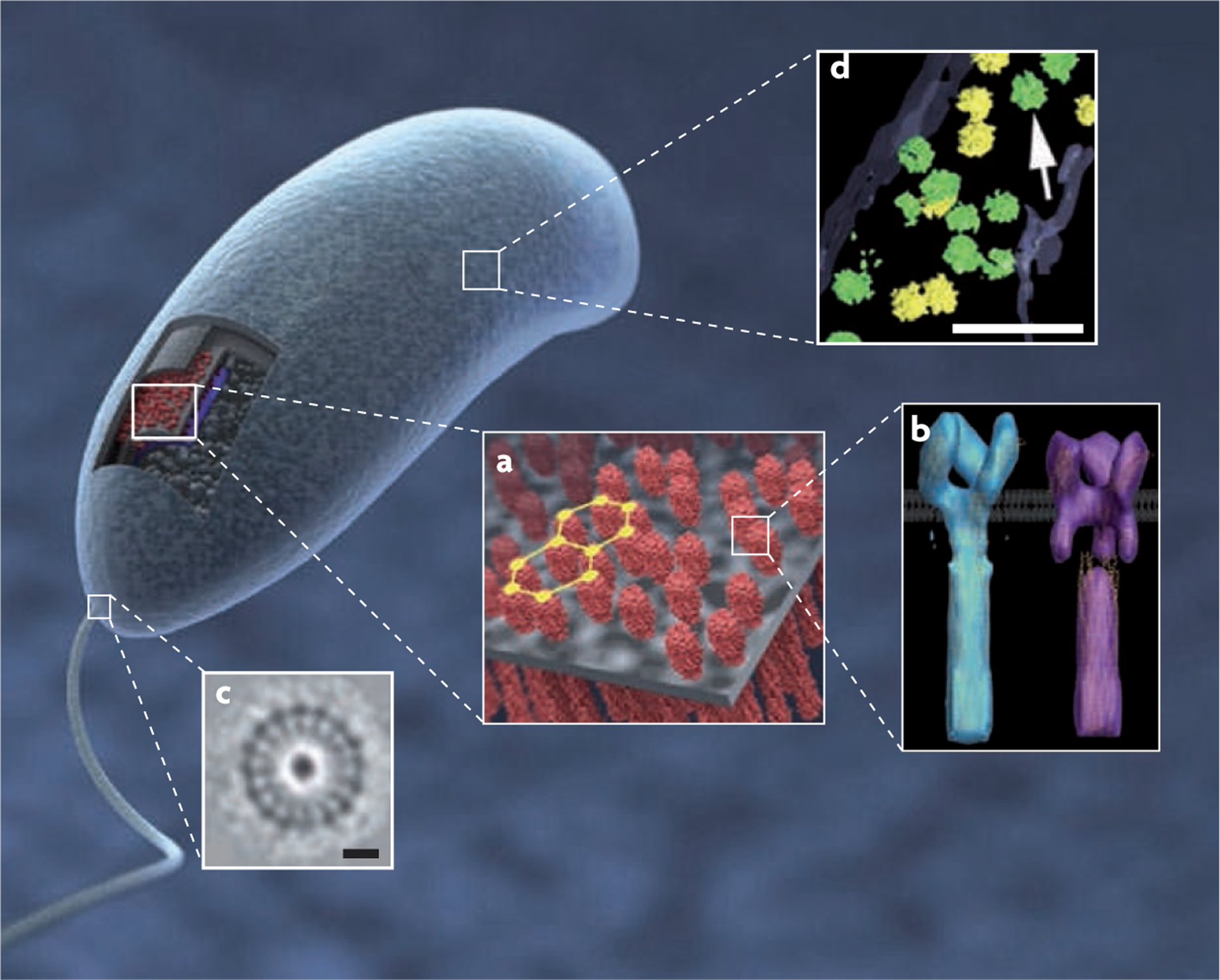
a | Visualization of the three-dimensional structure (3D) and partially ordered hexagonal arrangement of chemoreceptor arrays in Caulobacter crescentus cells. b | The two predominant receptor conformations of the Escherichia coli serine receptor Tsr derived by 3D averaging, corresponding to the ‘kinase-off’ and ‘kinase-on’ signalling states. c | The flagellar motor in cells of the Treponema primitia (scale bar 20 nm). d | Distribution and structure of ribosomes (indicated by the arrow) in Spiroplasma melliferum with green and yellow indicating higher or lower levels of accuracy of detection (scale bar 100 nm). Part a is reproduced, with permission, from REF. 45 © (2008) American Society for Microbiology. Part b is reproduced, with permission, from REF. 87 © (2008) National Academy of Sciences. Part c is reproduced, with permission, from Nature REF. 38 © (2006) Macmillan Publishers Ltd. All rights reserved. Part d is reproduced, with permission, from REF. 86 © (2006) Academic Press.
