Figure 1. Reconstitution of Cse4 Nucleosomes.
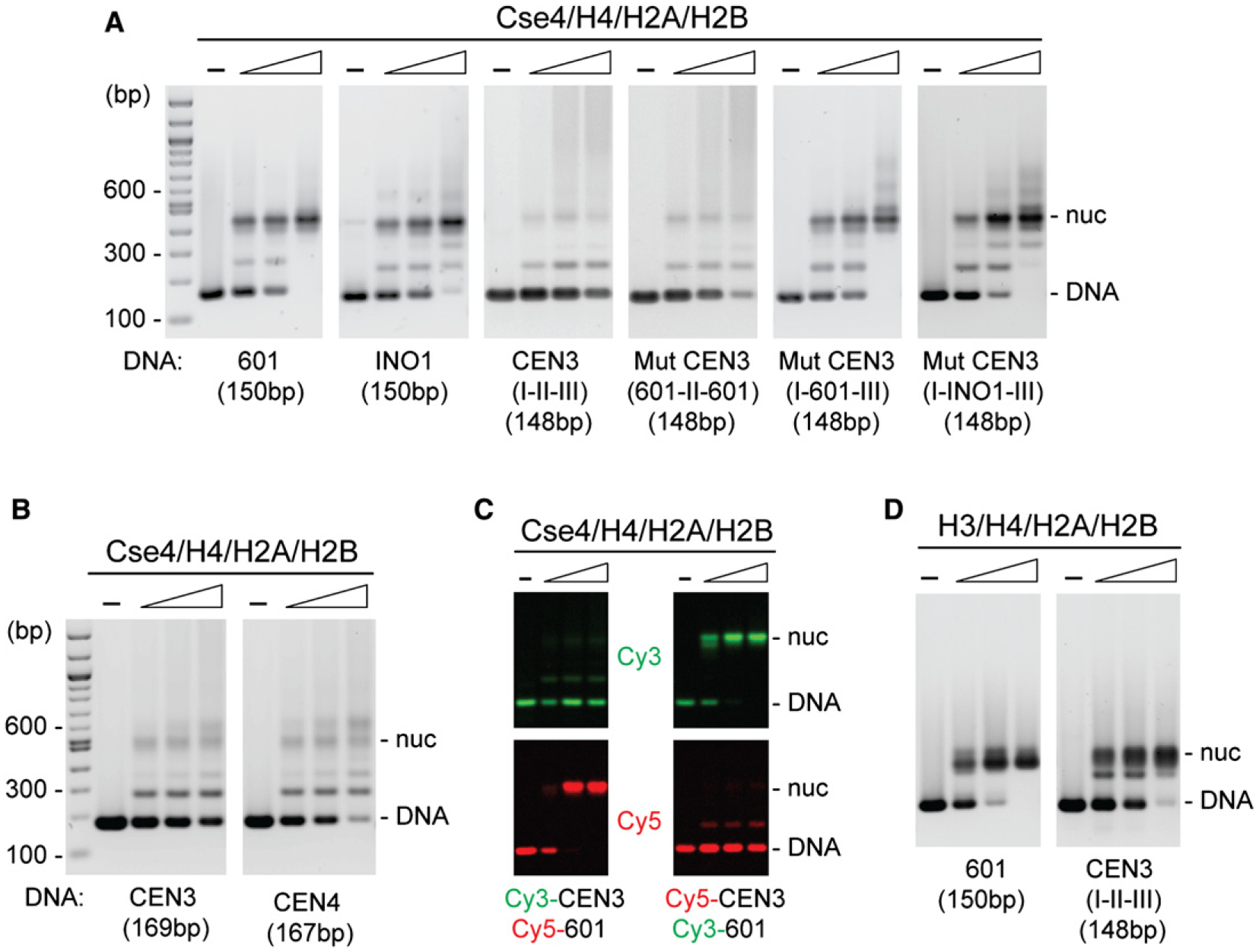
Octamers were mixed with DNA in 2 M NaCl, followed by overnight salt gradient dialysis at 4°C. Reconstituted nucleosomes were analyzed by EMSA and SYBR Green staining.
(A) Cse4 mononucleosomes were reconstituted using [Cse4/H4/H2A/H2B]2 octamers and the indicated DNAs: 150 bp 601, 150 bp INO1, 148 bp CEN3 (CDE I-II-III), and 148 bp mutant CEN3 (601-CDEII-601, CDE I-601-CDEIII, and CDE I-INO1-CDEIII) (protein:DNA molar ratio rm in ascending order, 0.3, 0.6, 1.2).
(B) CEN3 and CEN4 DNAs reconstitute Cse4 nucleosomes inefficiently.
(C) Competitive reconstitution of Cse4 nucleosomes with Cy3- and Cy5-labeled DNAs in the same reaction.
(D) Nucleosomes were reconstituted using [H3/H4/H2A/H2B]2 octamers and indicated DNAs (protein:DNA molar ratio rm in ascending order, 0.3, 0.6, 1.2). Samples were treated at 65°C overnight and analyzed by EMSA (see additional data in Figure S2).
