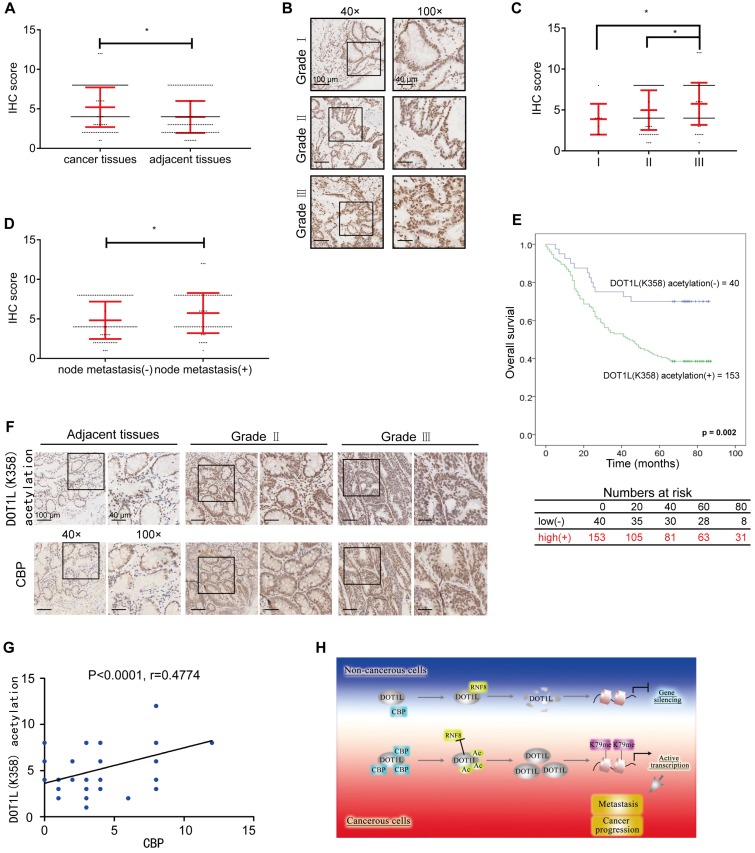
Figure 7.
DOT1L acetylation levels positively correlates with CBP expression and is associated with CRC metastasis and progression. (A) DOT1L(K358) acetylation staining on tissue microarrays (TMAs) containing 155 normal and CRC tissues. The relative DOT1L(K358) acetylation levels were compared between normal and CRC tissues (Student's t-test, n =155; the data represent the means ± SD, error bars are shown in red; *P<0.05). (B) IHC staining of TMAs containing 193 colon carcinoma samples (grades I, II, III) for DOT1L(K358) acetylation levels. Representative images of anti-DOT1L(K358) acetylation staining. Scale bars: 100 μm (left); 40 μm (right). (C) DOT1L(K358) acetylation staining scores were determined by evaluating the extent and intensity of immune-positivity and were analyzed by two-tailed unpaired student's t-test; the data represent the means ± SD, error bars are shown in red; *p < 0.05. (D) The relative DOT1L(K358) acetylation levels were compared between colon adenocarcinoma tissues with and without lymph node metastasis. The data were analyzed by Student's t-test, n =193; the data represent the means ± SD, error bars are shown in red; *P<0.05. (E) Kaplan-Meier curve showing the percentage overall survival of patients with colon cancer, stratified by DOT1L(K358) acetylation expression. (F) Samples from adjacent normal tissues and CRC cancers were immuno-stained with antibodies against DOT1L(K358) acetylation or CBP. Representative images are shown. Scale bars: 100 μm (left); 40 μm (right). (G) The CBP scores were plotted against the DOT1L(K358) acetylation scores, and the correlation coefficients were calculated by Pearson correlation analysis. (H) A proposed model of the DOT1L(K358) regulatory pathway in CRC carcinogenesis. Excessive CBP-mediated DOT1L acetylation in cancerous cells protects DOT1L from degradation via RNF8 and induces DOT1L enrichment. This effect enables H3K79 methylation of target genes and transcriptional activation of EMT transcription factors. The overall effect is promotion of tumorigenesis and metastasis.