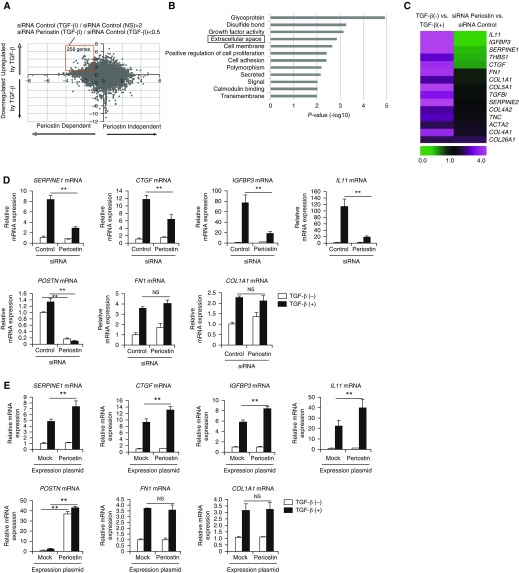
Figure 1.
Identification of signature molecules of pulmonary fibrosis induced by the cross-talk between TGF-β (transforming growth factor-β) and periostin in lung fibroblasts. (A) A dot plot of the genes showing dependency of either TGF-β or periostin analyzed by DNA microarray. We incubated normal human lung fibroblasts (NHLFs) with or without 1 ng/ml of TGF-β for 24 hours in the presence of 5 nM of either control siRNA or periostin siRNA, then subjected them to DNA microarray. The longitudinal axis represents the induction of the genes upregulated or downregulated by TGF-β. The horizontal axis represents the induction of the genes upregulated or downregulated by siRNA for periostin. The red square represents the genes upregulated by TGF-β by more than twofold and downregulated by periostin knockdown by less than half (258 genes). (B) Highly ranked Gene Ontology terms in the definite cross-talk genes between TGF-β and periostin in lung fibroblasts are depicted. “Extracellular space” is boxed. (C) Heat maps of the genes in the Gene Ontology term “extracellular space” are depicted. The ratios of control siRNA/TGF-β− to control siRNA/TGF-β+ (left) and control siRNA/TGF-β+ to periostin siRNA/TGF-β+ (right) are shown. (D) Effects of periostin knockdown on expression at mRNA levels of SERPINE1, CTGF, IGFBP3, IL11, POSTN, FN1, and COL1A1. NHLFs were stimulated with or without 1 ng/ml of TGF-β for 24 hours in the presence of control or periostin siRNA (n = 3). The values were adjusted by GAPDH expression. (E) MRC-5 cells were transiently transfected with the expression plasmid encoding periostin and then treated with or without 3 ng/ml of TGF-β for 24 hours. The mRNA expression of SERPINE1, CTGF, IGFBP3, IL11, POSTN, FN1, and COL1A1 is depicted. The values were adjusted by GAPDH expression. The same experiments were performed three times. **P < 0.01. NS = not significant.