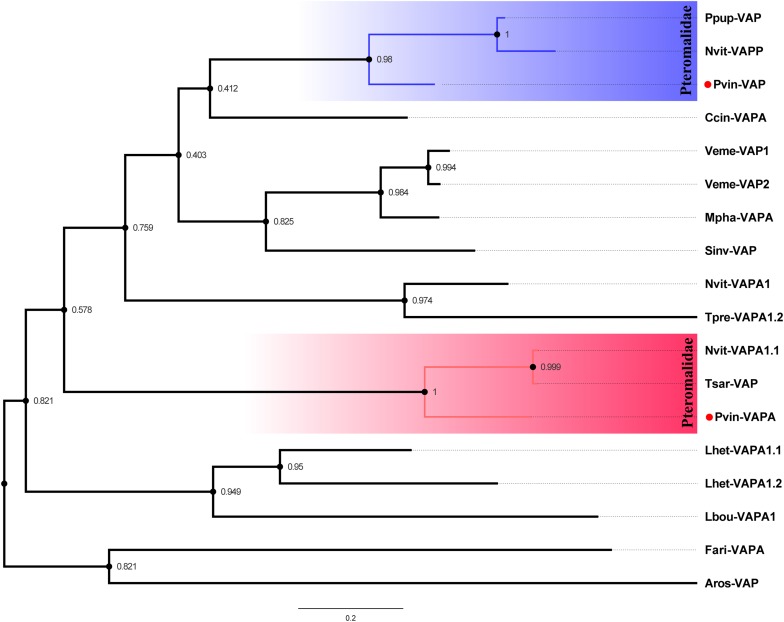
FIGURE 4.
Phylogenetic reconstruction based on “His_Phos_2 domain” of venom acid phosphatases. The phylogenetic tree was constructed by maximum likelihood method using the program Mega 6. 48 different amino acid substitution models were tested and the “LG+G+I” was considered to be the best model. The bootstrap values are presented on the nodes. Both venom proteins and non-venom orthologs were used for phylogenetic analysis. Full names of abbreviations are listed as follows. Pvin-VAP, P. vindemmiae venom acid phosphatase (Cluster-8535.24978); Pvin-VAPA, P. vindemmiae venom acid phosphatase Acph-1-like (Cluster-8535.6129); Ppup-VAP, P. puparum venom acid phosphatase (ACA60733.1); Nvit-VAPP, N. vitripennis venom acid phosphatase-like precursor (NP_001155147.1); Nvit-VAPA1.1, N. vitripennis venom acid phosphatase Acph-1-like (XP_001600770.1); Lhet-VAPA1.1, L. heterotoma venom acid phosphatase Acph-1-like1 (comp1442_c0_seq1); Lhet-VAPA1.2, L. heterotoma venom acid phosphatase Acph-1-like2 (comp2636_c0_seq1); Lbou-VAPA1, L. boulardi venom acid phosphatase Acph-1-like (comp9544_c0_seq1); Tpre-VAPA1.2, Trichogramma pretiosum venom acid phosphatase Acph-1-like isoform X1 (XP_014234174.2); Fari-VAPA, Fopius arisanus venom acid phosphatase Acph-1-like (XP_011310108.1); Tsar-VAP, T. sarcophagae venom acid phosphatase (OXU23470.1); Mpha-VAPA, Monomorium pharaonis venom acid phosphatase Acph-1-like (XP_012537166.1); Ccin-VAPA, Cephus cinctus venom acid phosphatase Acph-1 (XP_015589422.1); Sinv-VAP, Solenopsis invicta venom acid phosphatase (XP_025987662.1); Veme-VAP1, Vollenhovia emeryi venom acid phosphatase Acph-1-like isoform X1 (XP_011864393.1); Veme-VAP2, V. emeryi venom acid phosphatase Acph-1-like (XP_011872642.1); Aros-VAP, A. rosae venom acid phosphatase Acph-1-like (XP_012251812.1).