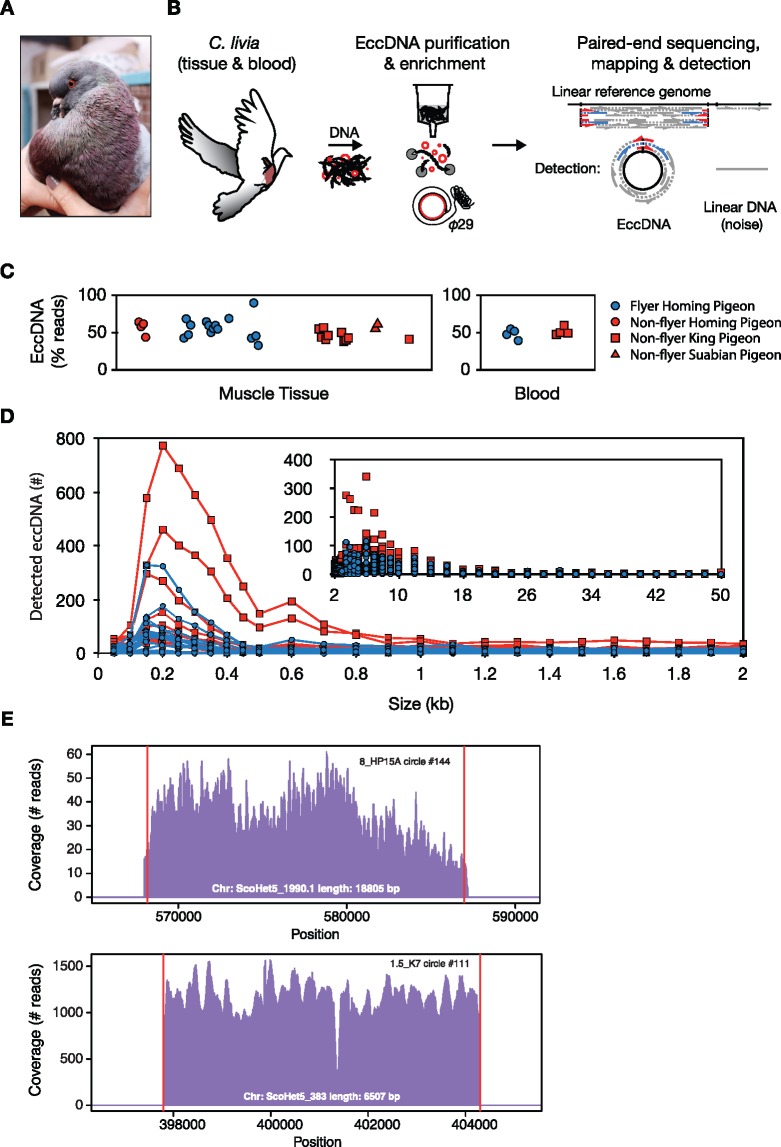
Fig. 1.
—EccDNA purification, detection, and size distribution in pigeon muscles and blood. (A) Picture of a king pigeon (Colombia livia). (B) Schematic description of the Circle-Seq method used for genome-wide profiling of eccDNA from pigeon, that is, eccDNA elution through a column ion-exchange membrane, removal of reminiscent linear DNA by exonuclease, rolling-circle amplification by phi29 polymerase, and finally paired-end sequencing, mapping, and detection of eccDNA from paired-end reads. EccDNAs were recorded with high confidence when at least two reads supported the circle junction (discordant reads, blue; soft-clipped or split reads, red) and read coverage was more than 80% from concordant read pairs, gray. (C) Read percentage on detected eccDNA relative to total mapped reads to the pigeon genome. x axis, sample separation by group and age in arbitrary units (not shown). (D) Length distribution of eccDNAs in muscle samples, each from ∼1E + 06 muscle nuclei (n = 27), grouped in bins of 50 bp (0–500 bp), 100 bp (500–3000 bp), 1,000 bp (3,000–50,000) intervals. (E) Representative read-coverage plots from a locus in homing pigeon (8_HP15A, upper plot) and a locus in king pigeon (1.5_K7, lower plot), detected as original sites of eccDNA formation.