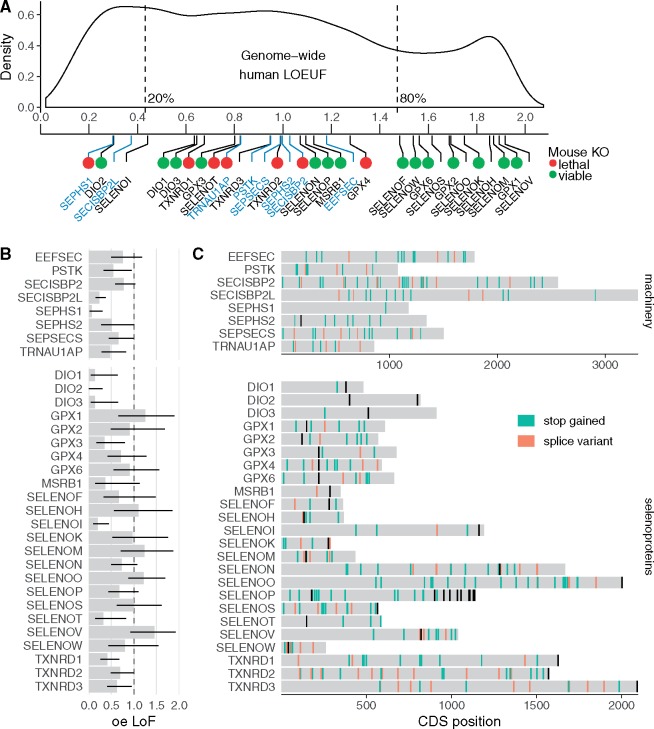
Fig. 1.
Loss-of-function (LoF) variants in human selenoproteins and tolerance to gene inactivation. (A) Genome-wide LOEUF score distribution of human genes and the corresponding values for selenoproteins (black labels) and Sec machinery genes (blue labels). A lower LOUEF indicates stronger selective constraints against LoF variants. Green and red dots indicate whether global homozygous deletion of the ortholog in mice is viable or lethal, respectively. IMPC reports contradictory phenotype for GPX3 and SECISBP2 (supplementary table S2, Supplementary Material online). (B) oe (observed/expected) LoF score for machinery genes and selenoproteins. Thin black lines correspond to the 90% confidence interval (its upper limit, LOEUF, is shown in panel A). (C) Observed nonsense and splice variants in human Sec machinery genes and selenoproteins (allele frequency <= 0.1 and median coverage >= 1) in the CDS of the representative transcripts (supplementary table S5, Supplementary Material online). Black sites correspond to annotated UGA-Sec. Variants and gene constraint scores from gnomAD v2.1. LOEUF, LoF oe upper-bound fraction (oe LoF 90% CI upper limit); oe LoF, observed/expected loss-of-function ratio; CDS, coding sequence.