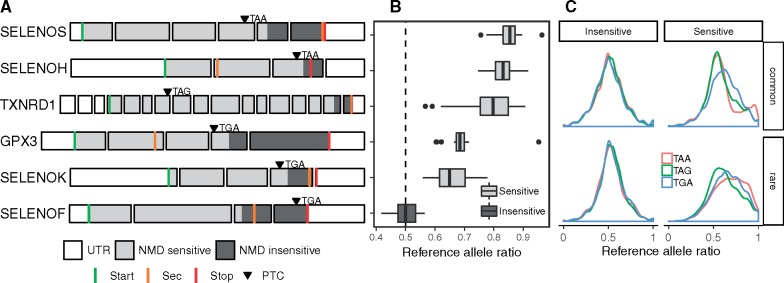
Fig. 3.
Presence of premature termination codon (PTC) in selenoprotein transcripts triggers their nonsense-mediated decay (NMD) in human tissues. (A) Exon structure of selenoprotein genes in which a PTC (shown by arrowheads) was identified in GTEx subjects. The corresponding stop codon is indicated. NMD sensitive and insensitive transcript regions (based on the 50-bp rule) are shaded in light and dark gray, respectively. (B) Allele-specific expression (ASE) of selenoprotein transcripts with PTC events. Distribution of the reference allele ratios (proportion of RNA-seq reads supporting the reference allele) observed across multiple tissues. Expression from both alleles results in 0.5 score (shown by the dashed vertical line). Imbalance toward 1 indicates depletion of the allele carrying the PTC. (C) Distribution of reference allele ratios in 2,456 genome-wide PTCs in GTEx subjects. PTCs are classified as insensitive or sensitive to NMD, and as common or rare based on allele frequency (threshold 0.01). UTR, untranslated region; Sec, selenocysteine UGA codon. Transcript length not at scale; only 100 nt of 3′UTR shown. PTC variants and ASE data obtained from GTEx v7.