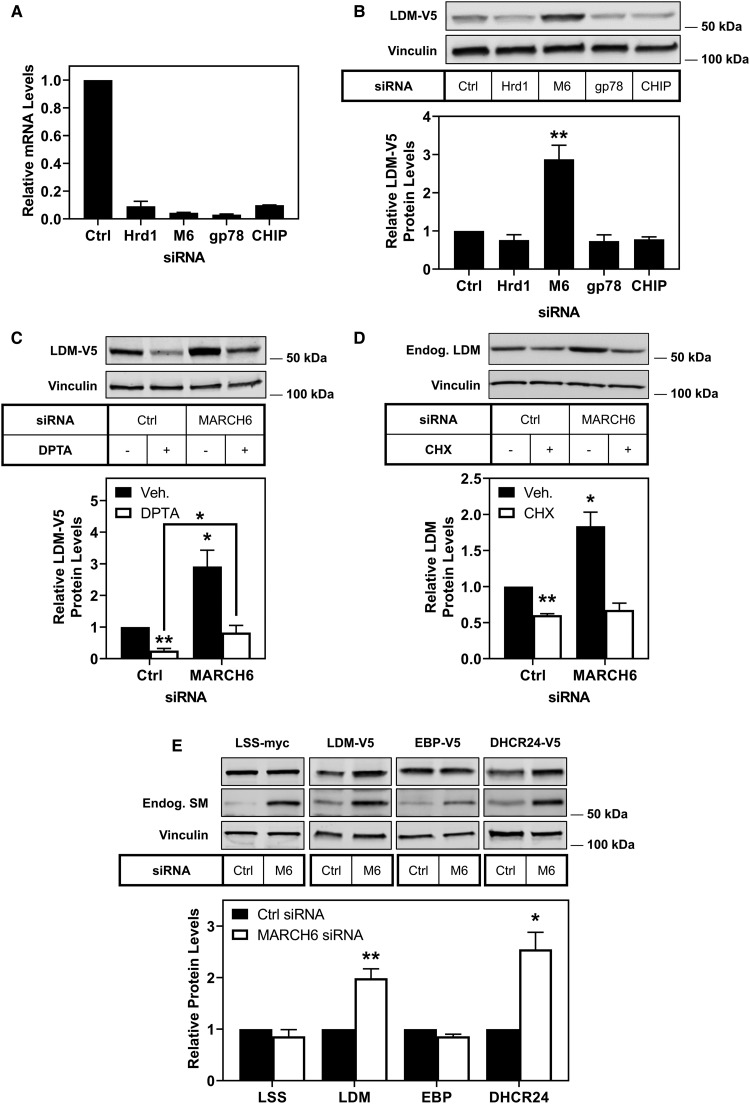
Figure 4. LDM and DHCR24 are degraded by the E3 ubiquitin ligase MARCH6.
(A) CHO-LDM-V5 cells were transfected for 24 h with 25 nM of the indicated siRNA (M6: MARCH6), then mRNA levels were measured using qRT-PCR and normalised to the housekeeping gene PBGD. mRNA levels were normalised to the control condition which was set to 1. Data are presented as mean + half range from an experiment performed in triplicate. (B) CHO-LDM-V5 cells were transfected for 24 h with 25 nM of the indicated siRNA (M6: MARCH6). (C) CHO-LDM-V5 cells were transfected for 24 h with 25 nM control or MARCH6 siRNA, then treated with or without 500 µM DPTA NONOate (DPTA) for 2 h. (D) HepG2 cells were transfected for 24 h with 25 nM control or MARCH6 siRNA, then treated with or without 10 µg/ml cycloheximide (CHX) for 8 h. (E) CHO-LSS-myc, CHO-LDM-V5, CHO-EBP-V5 or CHO-DHCR24-V5 cells were transfected for 24 h with 25 nM control or MARCH6 (M6) siRNA. Bands for the ectopic cholesterol synthesis enzymes migrated close to the expected size; LSS-myc: 83 kDa, LDM-V5: 57 kDa, EBP-V5: 26 kDa, DHCR24-V5: 60 kDa. Protein levels were analysed by Western blotting with V5, myc, endogenous SM, endogenous LDM and vinculin antibodies. Data are presented as mean + SEM from at least three independent experiments (B n = 3–7, C n = 6, D n = 3, E n = 3–6), where * P < 0.05 and ** P < 0.01. Relative protein levels were measured using ImageStudio Lite (version 5.2) and normalised to the control condition which was set to 1.