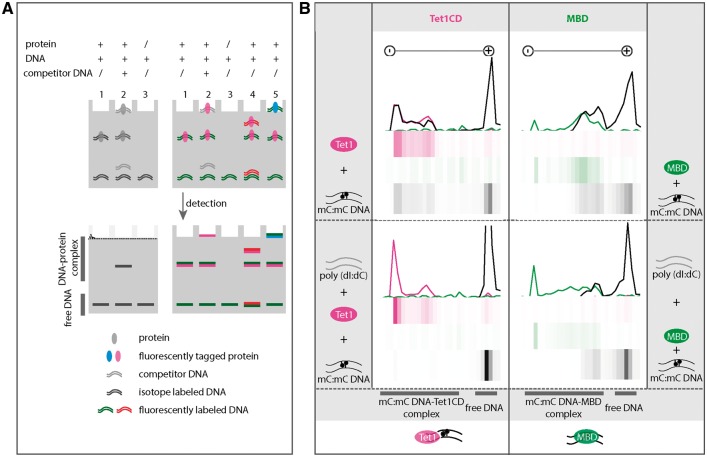
Figure 2:
Binding specificity of Tet1 to DNA. (A) Comparison of conventional EMSA (left, lanes 1–3) and multicolor EMSA (right lanes 1–5). Left lane 1: protein with isotope labeled DNA. Left lane 2: protein with isotope labeled DNA and competitor DNA. Left lane 3: isotope labeled DNA. Right lane 1: fluorescently tagged protein with fluorescently labeled DNA. Right lane 2: fluorescently tagged protein with fluorescently labeled DNA and competitor DNA. Right lane 3: fluorescently labeled DNA. Right lane 4: fluorescently tagged protein with two different fluorescently labeled DNAs. Right lane 5: Two different fluorescently tagged proteins with fluorescently labeled DNAs. (B) Binding specificity of Tet1CD (catalytic domain of Tet1) and MBD of Mecp2 (methyl-CpG binding protein 2) to methylated DNA. Purified mcherry-Tet1CD (2 µM) (magenta) was incubated with a 42-bp long oligonucleotide (1 µM) containing a symmetrically methylated CpG site (black) at 37 °C. After 90 min, free DNA and DNA-Tet1CD complexes were separated on a native polyacrylamide gel. As a control, the MBD protein (green, 2 µM) of Mecp2 was used. Minus and plus indicate cathode and anode, respectively. The fluorescence intensities were detected on a fluorescence microplate reader (see “Materials and methods” section) and intensities were plotted as line plots and heatmaps using RStudio. The upper part shows binding of Tet1CD (left) and the MBD of Mecp2 (right) to methylated DNA. The lower part shows binding of Tet1CD (left) or MBD (right) to methylated DNA in the presence of competitor DNA poly(dI:dC) (grey, 200 ng). Independent experiments were repeated twice. Shown is one representative result. A summary of the binding specificity of Tet1CD and MBD is shown at the bottom of the figure.