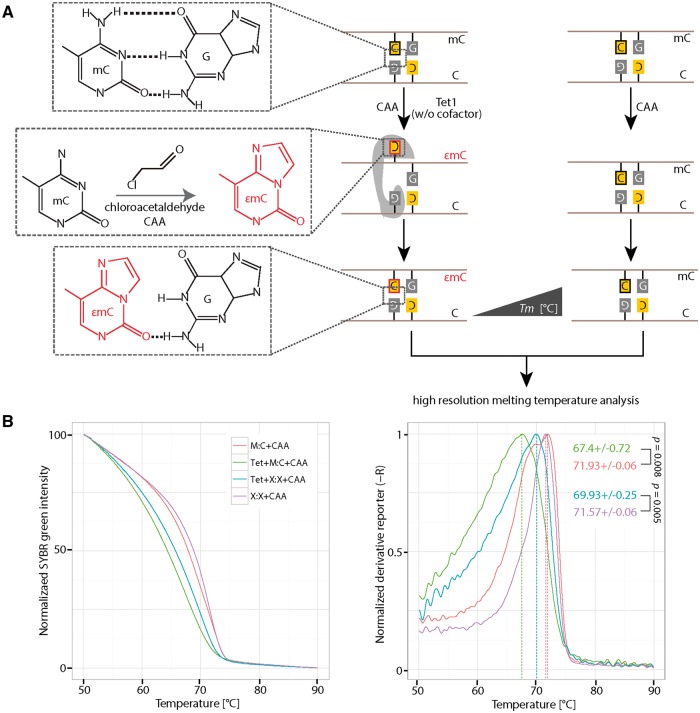
Figure 5:
Detection of Tet1 mediated DNA base flipping. (A) 0.5 µM of hemimethylated or non-CCGG containing DNA was incubated with or without 2 µM of the catalytic domain of Tet1 (Tet1CD) in the presence of 30 mM CAA. CAA reacts with 5mC when it is flipped out of the DNA helix. The reaction product 3,N4-ethenomethylcytosine (εmC) disrupts hydrogen bonds with base guanine in the complementary strand. The presence of 3,N4-ethenomethylcytosine (εmC) can be detected with HRM analysis, due to its low Tm contribution to double-stranded DNA. (B) Normalized fluorescent SYBR green intensities (left) and the corresponding derivative of intensities (right) were plotted using RStudio. Independent experiments were repeated two times. Shown is one representative result with three averaged technical replicates. The P-values of student’s t-test are indicated in the plot. Tm: melting temperature. C: cytosine; G: guanine; M:C: hemi methylated DNA; X:X: non-CpG containing DNA.