Abstract
It is widely accepted that CD4+ and CD8+ T-cells play a significant role in protection against Salmonella enterica serovar Typhi (S. Typhi), the causative agent of the typhoid fever. However, the antigen specificity of these T-cells remains largely unknown. Previously, we demonstrated the feasibility of using a recombinant Escherichia coli (E. coli) expression system to uncover the antigen specificity of CD4+ and CD8+ T cells. Here, we expanded these studies to include the evaluation of 12 additional S. Typhi proteins: 4 outer membrane proteins (OmpH, OmpL, OmpR, OmpX), 3 Vi-polysaccharide biosynthesis proteins (TviA, TviB, TviE), 3 cold shock proteins (CspA, CspB, CspC), and 2 conserved hypothetical proteins (Chp 1 and Chp2), all selected based on the bioinformatic analyses of the content of putative T-cell epitopes. CD4+ and CD8+ T cells from 15 adult volunteers, obtained before and 42 days after immunization with oral live attenuated Ty21a vaccine, were assessed for their functionality (i.e., production of cytokines and cytotoxic expression markers in response to stimulation with selected antigens) as measured by flow cytometry. Although volunteers differed on their T-cell antigen specificity, we observed T-cell immune responses against all S. Typhi proteins evaluated. These responses included 9 proteins, OmpH, OmpR, TviA, TviE, CspA, CspB, CspC, Chp 1 and Chp 2, which have not been previously reported to elicit T-cell responses. Interestingly, we also observed that, regardless of the protein, the functional patterns of the memory T-cells were different between CD4+ and CD8+ T cells. In sum, these studies demonstrated the feasibility of using bioinformatic analysis and the E. coli expressing system described here to uncover novel immunogenic T-cell proteins that could serve as potential targets for the production of protein-based vaccines.
Keywords: Salmonella, T-cells, recombinant E. coli, vaccine, human
1. Introduction
As for other intracellular infections, cellular-mediated immune responses against S. Typhi infection rely primarily on two types of cells: CD4+ and CD8+ T cells [1–4]. The presence of CD4+ helper T cells and classical class-Ia and non-classical-Ib restricted S. Typhi-specific CD8+ T cells have been observed in individuals with typhoid fever or immunized with Ty21a and other attenuated typhoid vaccine candidates [3, 5–16]. S. Typhi-specific CD8+ T cells have also been observed in humans challenged with wild-type S. Typhi [17, 18]. Typhoid vaccines have the potential to be cost-effective measures towards combating S. Typhi infection, yet the antigens triggering T-cell immune responses are largely unknown. Because humans are the only natural host for S. Typhi, and there is a lack of a suitable small animal model, few studies have investigated the T-cell immune responses to specific S. Typhi proteins in humans during infection and vaccination. Most of the S. Typhi proteins described as being involved in human protection have been derived from studies using mouse models of Salmonella infection [19, 20]. In the few studies using human samples, the investigators used peptide pools to evaluate T-cell responses, without protein processing by the antigen presenting cells. Indeed, using samples from individuals immunized with Ty21a typhoid vaccine, our group has previously demonstrated increases in the frequency of IFN-γ secreting CD8+ T cells in the presence of target cells coated with peptides that contain S. Typhi GroEL binding motifs [5]. Also, using peptide pools, a recent paper from Cerundolo’s group [21] have shown CD4+ T cells specific to Hemolysin E (HlyE) and cytolethal distending toxin B (CdtB), a component of typhoid toxin expressed by S. Typhi and S. Paratyphi A [22]. A downside of using peptide technology is the necessity that target cells express an HLA type able to bind the peptides and to ensure the individuals to be evaluated express the appropriate HLA alleles capable of presenting these antigens to T cells [23]. Using an innovative approach, i.e., microarray-based transcriptional analyses, Sheikh et al. demonstrated that the transcripts present in the blood of S. Paratyphi A naturally infected humans are expressed from PhoP and SlyA-regulated genes associated with intramacrophage survival, genes contained within Salmonella Pathogenicity Islands (SPIs) 1, as well as RpoS-regulated genes [24]. However, in this study, no immune responses against the expressed proteins were evaluated.
To overcome these limitations, our group recently has modified an antigen-expressing system, initially developed by the Higgins laboratory [25, 26] and based on the infection of B-cells with recombinant Escherichia coll (E. coli) to evaluate T-cell responses to four S. Typhi proteins: SifA, FliC, GroEL, and OmpC [27]. We found that all the tested individuals had increased T-cell responses over baseline (before immunization) to at least one of the four S. Typhi proteins evaluated. Moreover, multifunctional CD4+ and CD8+ T cells that expressed two or more cytokines, interleukin (IL)-17A, interferon (IFN-γ)-γ and tumor necrosis factor (TNF)-α), and/or CD107a/b molecules were detected [27]. These encouraging results prompt us to expand these studies to include the evaluation of 12 additional S. Typhi proteins: 4 outer membrane proteins (OmpH, OmpL, OmpR, and OmpX), 3 Vi-polysaccharide biosynthesis proteins (tviA, tviB, and tviE), 3 cold shock proteins (CspA, CspB, and CspC), and 2 conserved hypothetical proteins (Chp 1 and 2) (Table 1), all predicted to induce T-cell responses. These predictions were based on unique immunoinformatics tools developed by De Groot and Martin that systematically search for key determinants of immunity in available genome sequence data [28, 29].
Table 1.
S. Typhi proteins evaluated in this manuscript
| Protein Name |
Gene Name |
Accession Number |
Function |
MW (kd)** |
|---|---|---|---|---|
| Cold shock protein CspA | cspA | NP_807488 | Promote quick adaptation to temperature downshifts in the environment | 7.4 |
| Cold shock protein | cspB | NP_804712 | Function similar to CspA | 7.72 |
| Cold shock-like protein CspC | cspC | NP_804858 | Function similar to CspA | 7.4 |
| Conserved hypothetical protein 1* | chp1* | NP_805573 | NA | 5.84 |
| Conserved hypothetical protein 2* | chp2* | NP_807608 | NA | 35.3 |
| Outer membrane protein OmpH | ompH | NP_804107 | Unfolded protein biding | 17.9 |
| Porin OmpL | ompL | NP_807247 | Allows an efficient diffusion of low-molecular-weight solutes such as small sugars and tetraglycine | 27.13 |
| Transcriptional regulatory protein OmpR | ompR | NP_807614 | Required for the transcriptional expression of both major outer membrane protein genes OmpF and OmpC | 27.35 |
| Outer membrane protein X | ompX | NP_805818 | Promote bacterial adhesion to and entry into mammalian cells | 18.49 |
| Vi polysaccharide biosynthesis protein TviA | tviA | NP_807946 | Vi polysaccharide biosynthetic process | 21.11 |
| Vi polysaccharide biosynthesis protein VipA/TviB | tviB | NP_807945 | Function similar to TviA | 47.67 |
| Vi polysaccharide biosynthesis protein TviE | tviE | NP_807942 | Function similar to TviA | 64.98 |
NA, Not Available.
, Name not available. The name given in this manuscript is to facilitate the description of chp1 (tl800 locus) and chp2 (t3998 locus) gene results
, MW, Molecular weight obtained from The Universal Protein Resource (UniProt) database [https://www.uniprot.org/help/about]
We found that although volunteers differed in their T-cell antigen specificity, T-cell immune responses against all of the S. Typhi proteins that were evaluated were observed. Nine proteins that stimulated T-cell responses, OmpH, OmpR, TviA, TviE, CspA, CspB, CspC, Chp 1 and Chp 2, have not previously been reported in participants immunized with attenuated typhoid vaccines or exposed to wild-type S. Typhi. Interestingly, we also observed that, regardless of the protein, the functional patterns of the memory T-cells were different between CD4+ and CD8+ T cells. In sum, these studies confirmed and expanded the feasibility of using immunoinformatics analysis combined with the E. coli expressing system described here, to uncover novel immunogenic T-cell proteins that could serve as potential targets for the production of protein-based vaccines.
2. Methods
2.1. Ethics Statement
The clinical research was conducted following the human experimentation guidelines of the US Department of Health and Human Services and those of the University of Maryland, Baltimore that includes ethical standards laid down in the 1964 Declaration of Helsinki and the principles of the International Conference on Harmonization Good Clinical Practice guidelines [30]. All blood specimens were collected from volunteers who participated in the University of Maryland Institutional Review Board approved protocol number HP-00040022 that authorized the collection of blood samples from healthy volunteers for the studies included in this manuscript. The purpose and possible consequences of participating in this study were explained to the volunteers who gave informed, signed consent before the blood draws.
2.2. Participants
Fifteen healthy adult volunteers, aged 20-50 (37 ± 9) years, recruited from the Baltimore-Washington area and the University of Maryland at Baltimore campus, participated in this study. They were immunized with four spaced doses of 2-6 x 109 CFU of oral live attenuated Ty21a typhoid vaccine at an interval of 48 hours between doses [31, 32]. The blood was collected before and 42 days after Ty21a immunization. Peripheral blood mononuclear cells (PBMC) were isolated from their blood by density gradient centrifugation and cryopreserved in liquid N2 following standard techniques [6]. These PBMC were used ex vivo as effector cells or to prepare target cells.
2.3. Target cells
PBMC from Ty21a vaccinees obtained before immunization were used to generate autologous B-lymphoblastoid cell lines (B-LCL) to serve as target cells [5–8, 27, 33]. Briefly, supernatants from B95–8 cells (ATCC# CRL1612) containing the Epstein-Barr Virus (EBV) were used to infect and transform human B-lymphocyte-containing PBMC. After transformation, B-LCL were maintained in culture in RPMI 1640 (Gibco, Grand Island, New York) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, 50 μg/ml gentamicin, 2 mM L-glutamine, 2.5 mM sodium pyruvate, 10 mM HEPES buffer and 10% heat-inactivated fetal bovine serum (R10) or cryopreserved until used in the experiments.
2.4. Immunoinformatic Analysis and Protein Selection
iVAX toolkit is an ensemble of T-cell epitope mapping algorithms developed EpiVax Inc. and made available through the Institute for Immunology and Informatics [iCubed], University of Rhode Island. The EpiMatrix algorithm parses input protein sequences into overlapping 9- and 10-mer frames and screens each of the derived frames for amino acid sequence patterns indicative of HLA binding [28]. The Conservatrix algorithm can be used to identify 9- and 10-mer frames conserved within multiple input proteins or genomes. Herein, the iVAX toolkit was used to analyze S. Typhi proteins for highly conserved T cell epitopes that would be presented by prevalent HLA in human populations. Two complete genomes were submitted for analysis including the full wild-type S. Typhi strain Ty2 genome sequence (predicted to encode 4,318 protein-coding genes [GenBank Accession NC_004631]) and the full wild-type S. Typhi strain CT-18 genome sequence (predicted to encode 4,395 protein-coding genes [GenBank Accession NC_003198]) [34, 35]. The Conservatrix algorithm was used to identify 9-mer and 10-mer peptides present in both input genomes. Each conserved segment identified by Conservatrix was then submitted to the EpiMatrix algorithm and analyzed for binding potential with respect to a panel of eight common Class II alleles (9-mer frames only) and eleven available Class I alleles (9-mer and 10-mer frames). High scoring segments were then mapped back to their respective source proteins. Input proteins were then ranked by conserved Class I and Class II epitope content and 12 high scoring proteins were selected for further testing. They include: 4 outer membrane proteins (OmpH, OmpL, OmpR, OmpX), 3 Vi-polysaccharide biosynthesis proteins (TviA, TviB, TviE), 3 cold shock proteins (CspA, CspB, CspC), and 2 conserved hypothetical proteins (Chp 1 and Chp2).
2.5. Cloning of S. Typhi proteins from entry clones to pET-DEST-Hly into Recombinant E. coli
Gateway cloning of S. Typhi proteins from entry clones provided by the Institute of Allergy and Infectious Disease (NIAID)-funded Pathogen Functional Genomics Resource Center (PFGRC) was performed as previously described [27]. Briefly, entry clone plasmids carrying genes of interest were extracted using the QIA prep kit (Qiagen, Valencia, CA), the destination plasmid pET161-DEST-Hly was extracted with the HiSpeed plasmid preparation kit (Qiagen), and DNA concentration for both was measured using a NanoDrop instrument (Thermo Scientific, Waltham, MA). Each of the S. Typhi protein encoding genes was then transferred in vitro from its entry clone into the destination plasmid through Gateway LR reaction to generate a protein expression vector. We also cloned a short non-coding sequence called “pmark” that carried stop codons into pET161-DEST-Hly as a negative control of protein expression. Finally, each protein expression vector was transformed into One Shot BL21(DE3) competent E. coli (Invitrogen) by chemical transformation.
2.6. Infection of target cells by recombinant E. coli
Glycerol stocks of recombinant E. coli were streaked on LB agar with 100 μg/ml carbenicillin and incubated at 37°C overnight to obtain single colonies. A single colony was then cultured at 37 °C in 5 ml LB broth containing 100 μg/ml carbenicillin. After ~4h of incubation at 37°C (OD~0.4), the culture was induced for protein expression with 100 μM Isopropyl β-D-1-thiogalactopyranoside (IPTG) and incubated for an additional 2 h. Bacteria were then spun down at 4,500 rpm for 15 min and the supernatant discarded. Recombinant E. coli was used to infect the B-LCL as previously described [27]. We evaluated 12 recombinant E. coli strains expressing the following S. Typhi proteins: 4 outer membrane proteins (OmpH, OmpL, OmpR, OmpX), 3 Vi-polysaccharide biosynthesis proteins (TviA, TviB, TviE), 3 cold shock proteins (CspA, CspB, CspC), and 2 conserved hypothetical proteins (Chp 1 & Chp2). Briefly, target cells were infected by incubation in plain RPMI at 37°C for 2 hours with one of the 12 recombinant E. coli strains at a 1:30 multiplicity of infection (MOI). After incubation, cells were washed and incubated for an additional 16–18 hours in complete R10 containing gentamicin (100 μg/ml) to kill extracellular and/or to detach cell-bound bacteria. B-LCL were then gamma-irradiated (3,000 rads), surface stained with anti-CD45, a marker abundantly expressed on the surface of hematopoietic cells [36], and used as target cells. To confirm E. coli infection, aliquots of targets were surface stained with rabbit anti-E. coli antigen polyclonal antibody (1:1000, Abcam).
2.7. Monoclonal antibodies for surface and intracellular staining
PBMC were stained with monoclonal antibodies (mAbs) to CD69 (clone TPI-55–3) (Beckman-Coulter, Miami, FL), CD4 (clone RPA-T4), CD8 (clone HIT8a), CD107a and b (clones H4A3 and H4B4 respectively), IFN-γ (clone B27), TNF-α (clone MAb11) (BD Pharmingen, San Diego, CA, USA), CD14 (clone TuK4), CD19 (clone SJ25-C1), CD45 (clone H130) (Invitrogen), IL-17A (clone eBio64DEC17), CD62L (clone DREG-56) (eBioscience, San Diego, CA), and CD3 (clone OKT3), CD45RA (clone HI100) (Biolegend, San Diego, CA). Antibodies conjugated to the following fluorochromes were used in these studies: Fluorescein isothiocyanate (FITC), PE-Cy5.5, PE-Cy7, V450, Brilliant Violet (BV)570, BV605, BV650, Energy Coupled Dye or PE-Texas-Red conjugate (ECD), allophycocyanin (APC)-Alexa 700, APC-eFluor 780, and Quantum Dot (QD) 800.
2.8. Effector cells and co-cultures
Ex vivo PBMC from vaccinees collected before and 42 days after immunization with Ty21a typhoid vaccine were used as effectors as previously described [27]. Briefly, PBMC were co-cultured with autologous B-LCL cells expressing the Salmonella-Hly proteins at an effector to target cell ratio of 5:1 in the presence of mAbs to CD107a and CD107b. These mAbs were used to measure degranulation, a mechanism essential for the killing of S. infected targets by the cytotoxic T-cells [37]. PBMC were cultured with target cells exposed to Hly only or Staphylococcus enterotoxin B (SEB) (10 μg/ml, Sigma) were used as negative and positive controls, respectively. After ~2 hours, protein transport blockers, Monensin (1 μg/ml, Sigma) and brefeldin-A (BFA) (2 μg/ml, Sigma), were added to the co-culture. After an additional 16-18 hours (overnight) incubation, cells were harvested, stained with a dead-cell discriminator, yellow fluorescent viability dye (Yevid, Invitrogen)[38, 39], followed by surface staining with mAbs against surface antigens (CD3, CD4, CD8, CD14, CD19, CD45RA, and CD62L), and fixation and permeabilization with Fix & Perm cell buffers (Invitrogen, Carlsbad, CA). Cells were then stained intracellularly for IL-17A, IFN-γ, TNF-α, and CD69. Finally, cells were fixed with 1% paraformaldehyde and analyzed by flow cytometry on an LSR-II instrument (BD Biosciences). Data were analyzed with WinList 9.0 (Verity Software House, Topsham, ME). Single lymphocytes were gated based on forward scatter height vs. forward scatter area characteristics. A “dump” channel was used to eliminate dead cells (Yevid+) as well as macrophages/monocytes (CD14+), B lymphocytes (CD19+) and targets (CD45+) from the analysis. Additional gating on CD3, CD4, CD8, CD45RA, and CD62L was performed to identify cytokine-producing (IFN-γ, TNF-α and IL-17A), and CD107a/b expressing T-cell subsets and their memory status. Net responses were calculated by subtracting the number of positive events of the negative control (Hly only) from the experimental (Salmonella-Hly proteins). Functional responses were considered specific for S. Typhi if the differential in the number of positive and negative events between experimental (Salmonella-Hly proteins) and negative control (pmark-Hly only) cultures were significantly increased (P < 0.01) using Z-test. Volunteers were considered responders if the net responses from the PBMC collected 42 days after immunization were greater than 0.1 % from the net responses of PBMC collected before immunization [27]. Flow cytometry experiments were performed at the Flow Cytometry and Mass Cytometry Core Facility of the University of Maryland School of Medicine Center for Innovative Biomedical Resources (CIBR), Baltimore, Maryland.
2.9. Statistical analysis
All statistical tests were performed using Prism software (version 7, GraphPad Software, La Jolla, CA). Comparisons between two groups were carried out by paired Student’s t-tests. Pearson Product Moment Correlation tests were used to perform correlation analysis. P values <0.05 were considered significant.
3. Results
3.1. Presence of diverse S. Typhi specific-T cells in individuals immunized with the Ty21a-oral typhoid vaccine
Previous work from our group has demonstrated the feasibility of using a recombinant E. coli expressing system to uncover the antigen specificity of CD4+ and CD8+ T cells. Here, we expanded these studies to include the evaluation of 12 additional S. Typhi proteins: 4 outer membrane proteins (OmpH, OmpL, OmpR, and OmpX), 3 Vi-polysaccharide biosynthesis proteins (TviA, TviB, and TviE), 3 cold shock proteins (CspA, CspB, and CspC), and 2 conserved hypothetical proteins (Chp 1 and 2)(Table 1). These proteins were selected based on the bioinformatic analyses of their putative T-cell epitope content as described in Materials and Methods. Ex-vivo PBMC from 15 Ty21a-immunized volunteers obtained before and 42 days after immunization were exposed to autologous B-LCL infected with recombinant E. coli expressing Hly only or co-expressing one of the 12 Salmonella gene products. After stimulation, their expression of IL-17A, IFN-γ and TNF-α cytokines and/or CD107a and b molecules was evaluated by flow cytometry. Effector cells stimulated by B-LCL infected with recombinant E. coli expressing Hly only or Staphylococcus enterotoxin B (SEB) were used as negative and positive controls, respectively. In agreement with our previous work [27], we observed that although volunteers differed in their T-cell antigen specificity, T-cell immune responses were observed against all S. Typhi proteins evaluated. These included 9 proteins that had not been previously described, OmpH, OmpR, TviA, TviE, CspA, CspB, CspC, Chp 1 and Chp 2. The pattern of these responses was variable, with some individuals responding to up to eleven proteins while others responded to only one or two proteins. Of note, one individual was unresponsive to all proteins studied. A summary of the CD4+ and CD8+T-cell responses to individual S. Typhi proteins are presented in Tables 2 and 3. To facilitate the analyses, we next divided the frequency of responders into two groups: (a) frequency <25% and (b) frequency >25%. We found that except for TviA and TviE, the overall frequencies of CD8+ T-cell responding to one specific S. Typhi protein was higher or equal than the frequencies of CD4+ T-cells. We also found that while the highest frequencies of CD4+immune responses were directed toward Omp and Tvi, the highest frequencies of CD8+ immune responses were directed toward Omp and Csp protein families. Interestingly, the profile of the immune responses differed between CD4+ and CD8+T-cells (Fig. 1 and Supplementary Fig. 1). Regardless of the S. Typhi protein being evaluated, CD8+ T-cells consistently expressed IFN-γ and CD107 a/b at a higher magnitude than CD4+ T-cells (p< 0.0001). In contrast, CD4+ T-cells expressed IL-17A at higher level than CD8+T-cells (p< 0.0001) (Fig. 1 and Supplementary Fig. 1A). No significant differences were observed between the magnitude of the immune responses of CD4+ and CD8+T-cells expressing TNF-α, which was very variable among the volunteers. Representative responses from selected volunteers are shown in Figs 2 and 3. In parallel analyses, we compared the expression levels between paired CD4 and CD8 responses. The correlation was very high between pairs expressing IL-17A, IFN-γ and TNF-α (Supplementary Fig. 1B). No correlation was found between pairs expressing CD107 a/b (Supplementary Fig. 1B). Thus, these results suggest that upon stimulation by S. Typhi antigens the host mounts a coordinated and complementary CD4+ and CD8+ T-cell responses to amplify the immune response and fight the pathogen.
Table 2.
Positive CD4+ T-cell responses to individual S. Typhi proteins.
| Statistic | Protein Secretion | CspA | CspB | CspC | Chp1 | Chp2 | OmpH | OmpL | OmpR | OmpX | TviA | TviB | TviE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total N (N responders*) | 13 (3) | 14 (3) | 14 (6) | 13 (1) | 13 (1) | 15 (5) | 15 (6) | 15 (6) | 15 (5) | 15 (7) | 15 (4) | 15 (6) | |
| % responders | 23.1 | 21.4 | 42.8 | 7.7 | 7.7 | 33.3 | 40.0 | 40.0 | 33.3 | 46.7 | 26.7 | 40.0 | |
| Responders % (N) | IL-17A | 0.0 (0) | 7.1 (1) | 7.1 (1) | 0.0 (0) | 0.0 (0) | 20.0 (3) | 20.0 (3) | 13.3 (2) | 20.0 (3) | 13.3 (2) | 6.7 (1) | 6.7 (1) |
| IFN-γ | 0.0 (0) | 7.1 (1) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 6.7 (1) | 13.3 (2) | 13.3 (2) | 13.3 (2) | 6.7 (1) | 13.3 (2) | 13.3 (2) | |
| TNF-α | 23.1 (3) | 21.4 (3) | 14.3 (2) | 0.0 (0) | 7.7 (1) | 26.7 (4) | 26.7 (4) | 33.3 (5) | 20.0 (3) | 20.0 (3) | 13.3 (2) | 20.0 (3) | |
| CD107a/b | 23.1 (3) | 7.1 (1) | 35.7 (5) | 7.7 (1) | 7.7 (1) | 20.0 (3) | 20.0 (3) | 6.7 (1) | 20.0 (3) | 20.0 (3) | 20.0 (3) | 20.0 (3) | |
, Functional responses were considered specific for S. Typhi if the differential in the number of positive and negative events between experimental (Salmonella-Hly proteins) and negative control (Hly only) cultures were significantly increased (P < 0.01) using Z-test. Volunteers were considered responders if the net responses from the PBMC collected 42 days after immunization were greater than 0.1 % from the net responses of PBMC collected before immunization.
Bold fonts represent frequency of responders to a specific protein above 25%.
Table 3.
Positive CD8+ T-cell responses to individual S. Typhi proteins.
| Statistic | Protein Secretion | CspA | CspB | CspC | Chp1 | Chp2 | OmpH | OmpL | OmpR | OmpX | TviA | TviB | TviE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total N (N responders*) | 13 (7) | 14 (7) | 14 (7) | 13 (6) | 13 (3) | 15 (7) | 15 (9) | 15 (6) | 15 (7) | 15 (4) | 15 (4) | 15 (4) | |
| % responders | 53.8 | 50.0 | 50.0 | 46.1 | 23.1 | 46.7 | 60.0 | 40.0 | 46.7 | 26.7 | 26.7 | 26.7 | |
| Responders % (N) | IL-17A | 0.0 (0) | 7.1 (1) | 14.3 (2) | 15.4 (2) | 7.7 (1) | 6.7 (1) | 13.3 (2) | 6.7 (1) | 6.7 (1) | 6.7 (1) | 13.3 (2) | 6.7 (1) |
| IFN-γ | 38.5 (5) | 28.6 (4) | 21.4 (3) | 30.8 (4) | 23.1 (3) | 26.7 (4) | 33.3 (5) | 20.0 (3) | 33.3 (5) | 26.7 (4) | 20.0 (3) | 20.0 (3) | |
| TNF-α | 46.2 (6) | 35.7 (5) | 21.4 (3) | 23.1 (3) | 7.7 (1) | 26.7 (4) | 40.0 (6) | 20.0 (3) | 40.0 (6) | 20.0 (3) | 13.3 (2) | 20.0 (3) | |
| CD107a/b | 7.7 (1) | 42.9 (6) | 7.1 (1) | 7.7 (1) | 0.0 (0) | 33.3 (5) | 53.3 (8) | 13.3 (2) | 20.0 (3) | 6.7 (1) | 6.7 (1) | 6.7 (1) | |
, Functional responses were considered specific for S. Typhi if the differential in the number of positive and negative events between experimental (Salmonella-Hly proteins) and negative control (Hly only) cultures were significantly increased (P < 0.01) using Z-test. Volunteers were considered responders if the net responses from the PBMC collected 42 days after immunization were greater than 0.1 % from the net responses of PBMC collected before immunization.
Bold fonts represent frequency of responders to a specific protein above 25%.
Fig. 1. Antigen Presentation of S. Typhi proteins by targets infected with recombinant E. coli.
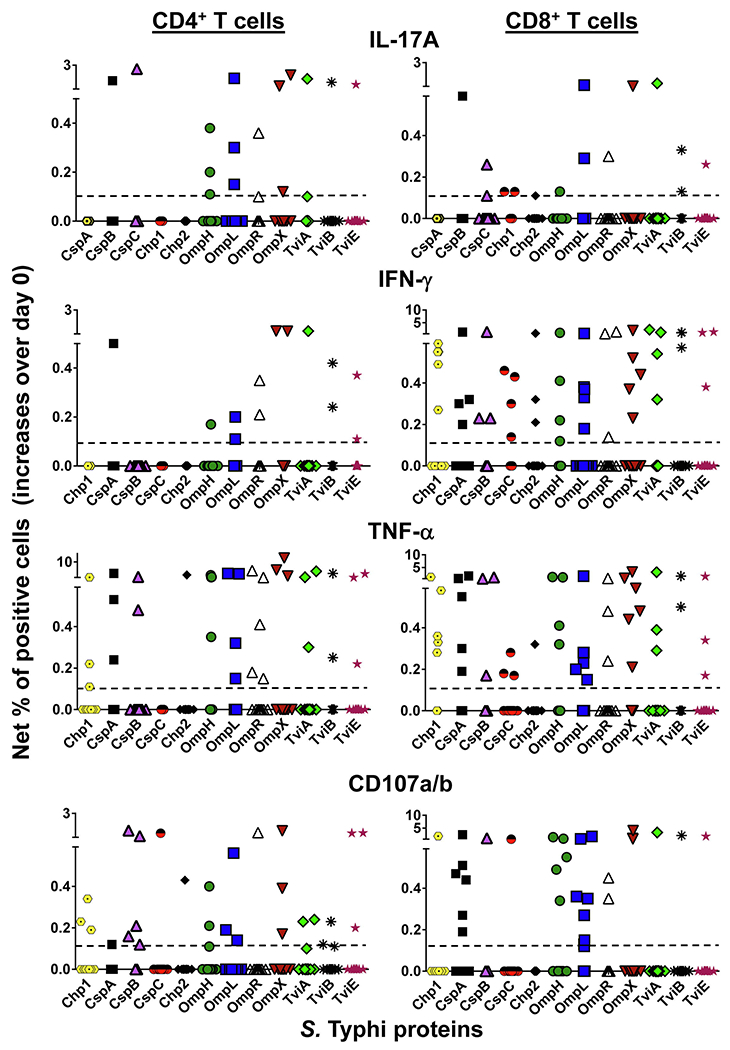
Ex vivo PBMC from 15 volunteers collected before and 42 days after immunization were co-cultured for 16–18 hrs with autologous B-LCL targets infected at 1:30 MOI with recombinant E. coli expressing Hly only or co-expressing S. Typhi antigens: CspA, CspB, OmpH, OmpL, OmpR, OmpX, TviA, TviB, TviE, and two conserved hypothetical proteins (Chp 1 and Chp2). After incubation cells were stained, and the ability of PBMC to produce one or more cytokines (IL-17A, IFN-γ and TNF-α) and/or express CD107a/b molecules was analyzed by flow cytometry. Two T-cell subset responses (i.e., CD4+ and CD8+ T cells) were evaluated. Net responses were calculated by subtracting the T-cell responses to B-LCLs infected with recombinant E. coli expressing S. Typhi/Hly antigens from the responses of the controls (B-LCL expressing Hly only). Increases over day 0 were calculated by subtracting the net responses of the PBMC collected before immunization from the net responses of PBMC collected 42 days after immunization. The dashed line represents the threshold for a positive response. The data represent the CD4+ and CD8+ T-cell responses of all 15 volunteers. Each colored symbol represents a distinct S. Typhi protein.
Fig. 2. CD4+ T-cell responses to S. Typhi proteins presented by targets infected with recombinant E. coli.
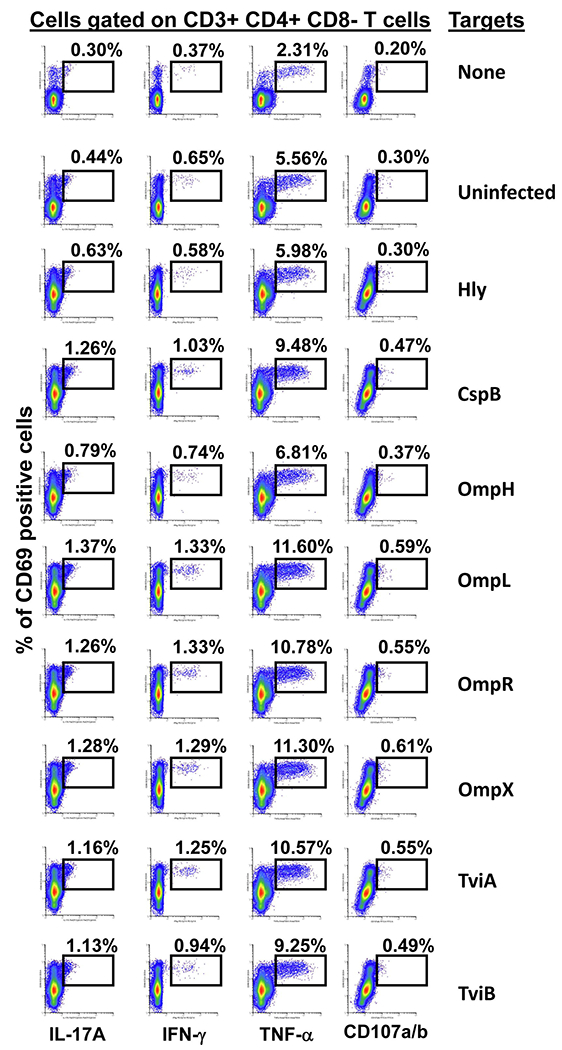
Ex vivo PBMC from a volunteer collected 42 days after immunization were co-cultured for 16-18 hrs with autologous B-LCL targets infected at a 1:30 MOI with one of the eight recombinant E. coli expressing S. Typhi/Hly (CspB, OmpH, OmpL, OmpR, OmpX, TviA and TviB) or only Hly (control) proteins. After incubation, cells were stained and the ability of the PBMC to produce one or more cytokines (IL-17A, IFN-γ and TNF-α) and/or express CD107a/b molecules was evaluated by flow cytometry. Shown are the CD4+ T-cell responses from a representative volunteer. Numbers represent the percentage of positive cells.
Fig. 3. CD8+ T cell responses to S. Typhi proteins presented by targets infected with recombinant E. coli.
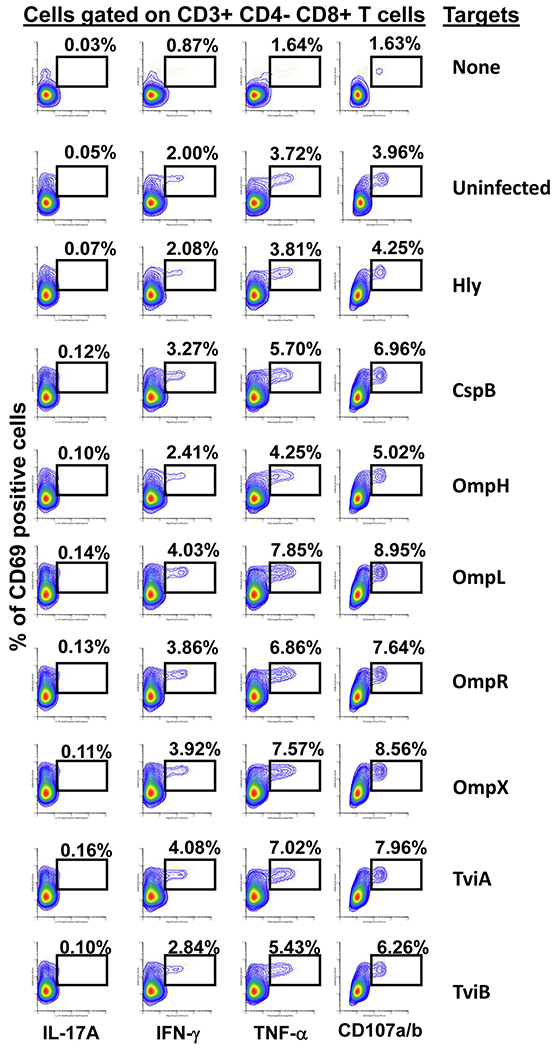
Ex vivo PBMC from a volunteer collected 42 days after immunization were co-cultured for 16–18 hrs with autologous B-LCL targets infected at a 1:30 MOI with one of the eight recombinant E. coli expressing S. Typhi/Hly (CspB, OmpH, OmpL, OmpR, OmpX, TviA and TviB) or only Hly (control) proteins. After incubation, cells were stained, and the ability of the PBMC to produce one or more cytokines (IL-17A, IFN-γ and TNF-α) and/or express CD107a/b molecules was evaluated by flow cytometry. Shown are the CD8+ T-cell responses from a representative volunteer. Numbers represent the percentage of positive cells.
3.2. Functionality of CD4+ T-cells and CD8+ T- cells
Previous results from our group have shown that multifunctional T-cells might contribute to S. Typhi immunity and protection [8, 13, 17, 27]. We therefore investigated the multi-functionality patterns of CD4+ and CD8+ T-cells after exposure to infected B-LCL infected with recombinant E. coli expressing S. Typhi proteins. We measured four T-cell functions simultaneously (i.e., production of IL-17A, IFN-γ and TNF-α cytokines, or expression of CD107a/b molecules) by multichromatic flow cytometry using the FCOM feature of the WinList software, which provides the % of T-cells expressing one of the 15 possible cytokine/CD107a/b combinations (i.e., single, double, triple or quadruple functions). Analyses of the frequency of single, double, triple or quadruple expressing cells revealed differences between the functional phenotypes of CD4+ and CD8+ T-cells. However, these expression patterns were consistent among the same subtype and did not vary in function of the protein being expressed by the target cells (Figs. 4 and 5). Single cytokine producers constituted the majority of CD4+ T-cell responses (Fig. 4). The most frequently detected phenotypes on CD4+ T-cells included single producers expressing either TNF-α (range, 52.5 to 70%) or IL-17A (range, 4 to 23%). These two phenotypes accounted for an average of 75% of the total CD4+ T-cell responses. The remaining CD4+ T-cells were single producers expressing CD107a/b (range, 1.5 to 8.5%) or double producers expressing either IL-17A and TNF-α (range, 1 to 9%) or IFN-γ and TNF-α (range, 6.5 to 11%) (Fig. 4). In contrast, the frequency of cells expressing triple functions was higher in CD8+ T-cells than in CD4+ T-cells. The most prominent CD8+ T-cell phenotype was among cells with concomitant expression of IFN-γ, TNF-α and CD107a/b (mean, 31.4 [range, 28 to 34.5%)(Fig. 5). CD8+ T-cells expressing double and single functions accounted for 27.2 and 39.9% of the total cells (Fig. 5), respectively. Interestingly, in most of the individuals studied, regardless of the T-cell subset, CD4+ or CD8+ T-cells, the concomitant expression of IFN-γ and IL-17A were at a very low level or absent (Figs. 4 and 5). Thus, it appears that the characteristics of the T-cell responses to S. Typhi proteins depends on the subset being examined (i.e., CD4+ or CD8+), and was only weakly affected (if at all) by the proteins being examined in this study.
Fig. 4. Frequencies of mono and multifunctional CD4+ T-cells.
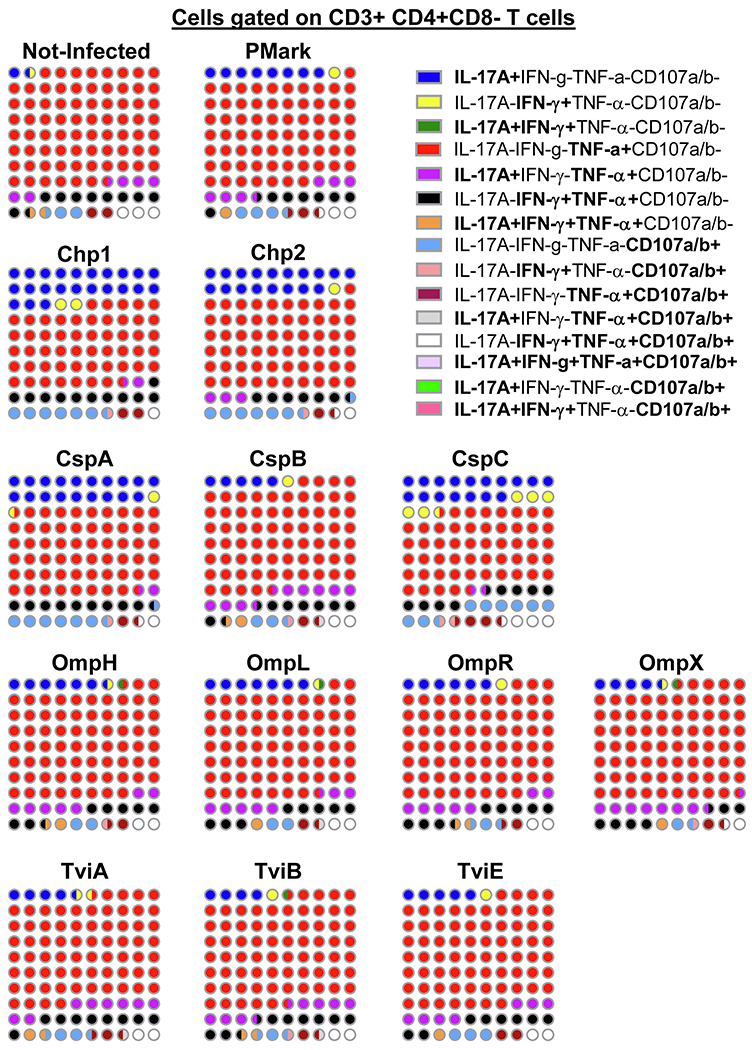
FCOM, an analysis tool contained in the WinList software package, was used to automatically reduce multiparameter data to a series of multiple event acquisition gates, one for each of the 15 possible sub-phenotypes of CD4+ T-cell multifunctionality. Lymphocytes were gated-out based on forward scatter height vs. forward scatter area. A “dump” channel was used to eliminate dead cells (Yevid+) as well as macrophages/monocytes (CD14+), B lymphocytes (CD19+) and targets (CD45+) from the analysis. This was followed by additional gating on CD3 and CD4 to identify single and multifunctional CD4+ T-cells. Combinations with frequency values that were zero were ignored. The data represents an average of 4 volunteers with sufficient number of positive events to allow for a detailed analysis of the T-memory subsets of the responding populations. Data presented as 10X10 matrixes in which each circle represents 1% of the population, color-coded as described in the legend. Bi-colored circles represent 0.5%.
Fig. 5. The frequency of mono and multifunctional CD8+ cells.

FCOM, an analysis tool contained in the WinList software package, was used to automatically reduce multiparameter data to a series of multiple event acquisition gates, one for each of the 15 possible sub-phenotypes of CD8+ T-cell multifunctionality. Lymphocytes were gated-out based on forward scatter height vs. forward scatter area. A “dump” channel was used to eliminate dead cells (Yevid+) as well as macrophages/monocytes (CD14+), B lymphocytes (CD19+) and targets (CD45+) from the analysis. This was followed by additional gating on CD3 and CD8 to identify single and multifunctional CD8+ T-cells. Combinations with frequency values that were zero were ignored. The data represents an average of 4 volunteers with sufficient number of positive events to allow for a detailed analysis of the T-memory subsets of the responding populations. Data presented as 10X10 matrixes in which each circle represents 1% of the population, color-coded as described in the legend. Bi-colored circles represent 0.5%.
3.3. Memory subset analyses of mono and multifunctional T-cells
We next used CD45RA and CD62L markers to identify the memory status of the mono and multifunctional T-cells by flow cytometry. To this end, subsequent gating was performed on the 15 functional phenotypes of T-cell subsets to identify 4 subpopulations: central memory (CD45RA-CD62L+, TCM), naive (CD45RA+CD62L+, Naive), effector memory (CD45RA-CD62L−, TEM), and effector memory expressing CD45RA (CD45RA+CD62L−, TEMRA). We evaluated the frequencies of the top 4 and 5 functional phenotypes among CD4+ and CD8+ T-cells, respectively. We found that naive CD4+ cells were enriched in cells capable of expressing IL-17A. Interestingly, while CD4+ cells with concomitant expression of IL-17A and TNF-α were present in high proportions of TEM and TEMRA cells, CD4+ cells with concomitant expression of IFN-γ and TNF-α were highly present largely in TEM cells (Fig. 6). Of note, TCM, although not the predominant subset, was present in considerable proportions of CD4+ cells expressing TNF-α alone or together with IL-17A (Fig. 6). In contrast, the proportion of TEM among CD8+ T cells increased as these CD8+ T cells became more multifunctional (Fig. 7). Indeed, the majority of mono-functional CD8+ T cells were distributed among TEM and TEMRA cells. Low frequencies of naïve and TCM were observed among CD8+ T cells. Finally, regardless of the T-cell subset evaluated, the TEMRA populations were enhanced in mono-functional cells. Representative responses from selected volunteers are shown in Figs 6 and 7. Together, these data indicate that CD4+ or CD8+ T-cells are heterogeneous in terms of their memory phenotype and functional properties. These phenotypes may have complementary capabilities in protective immune responses against S. Typhi.
Fig. 6. Memory status of the mono and multifunctional CD4+ T-cells.
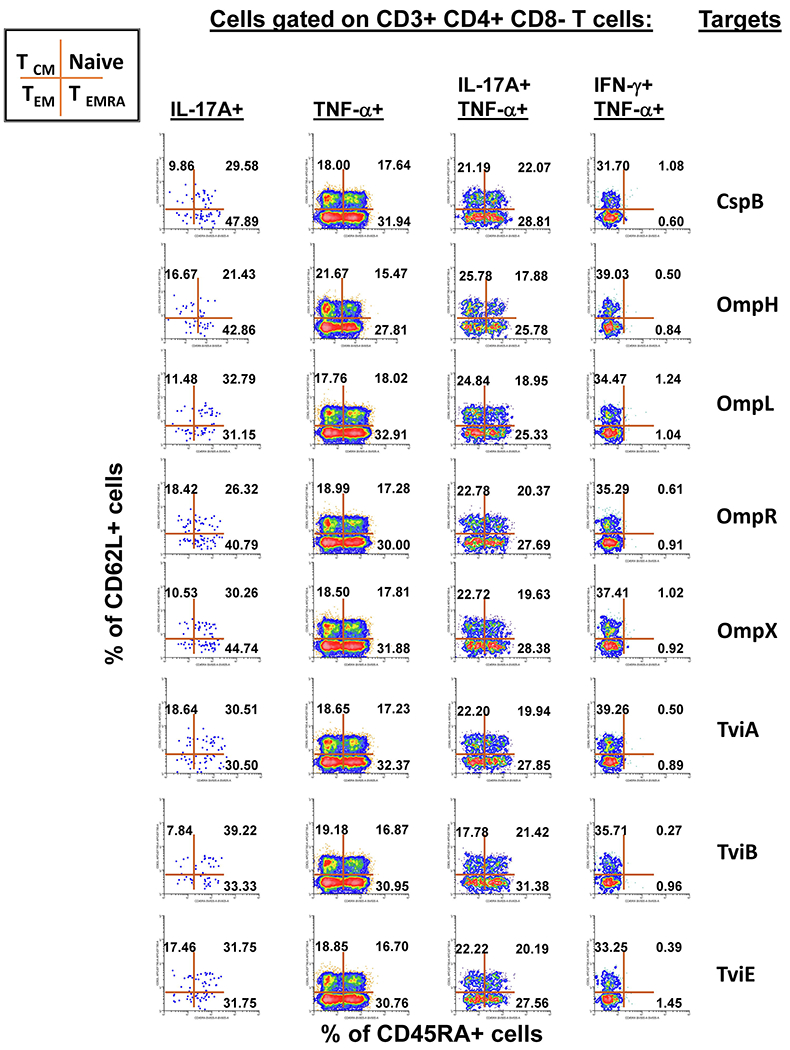
Subsequent gates on the mono and multifunctional CD4+ T-cell subsets described in Fig. 4 were used to identify the memory status among each of the 15 defined sub-phenotypes of CD4+ cells. The data is representative of one experiment/one volunteer showing mono-functional or multi-functional CD4+ T-cell responses to eight recombinant E. coli expressing S. Typhi/Hly (CspB, OmpH, OmpL, OmpR, OmpX, TviA, TviB, and TviE) proteins at a multiplicity of infection (MOI) of 1:30. This figure illustrates a gating strategy in which CD4+ T-cells are further categorized based on the expression of CD45RA and CD62L markers. Cells in each resulting quadrant of the dot plot are then categorized in 4 subpopulations: central memory (CD45RA−CD62L+, TCM), naive (CD45RA+CD62L+, Naive), effector memory (CD45RA−CD62L−, TEM), effector memory expressing CD45RA (CD45RA+CD62L−, TEMRA). The 4 selected populations are those that were dominant in the volunteers who responded to stimulation with S. Typhi proteins, exhibiting sufficient number of positive events to allow downstream analyses. Numbers represent the percentage of positive cells in the respective quadrant.
Fig. 7. Memory status of the mono and multifunctional CD8+ T-cells.
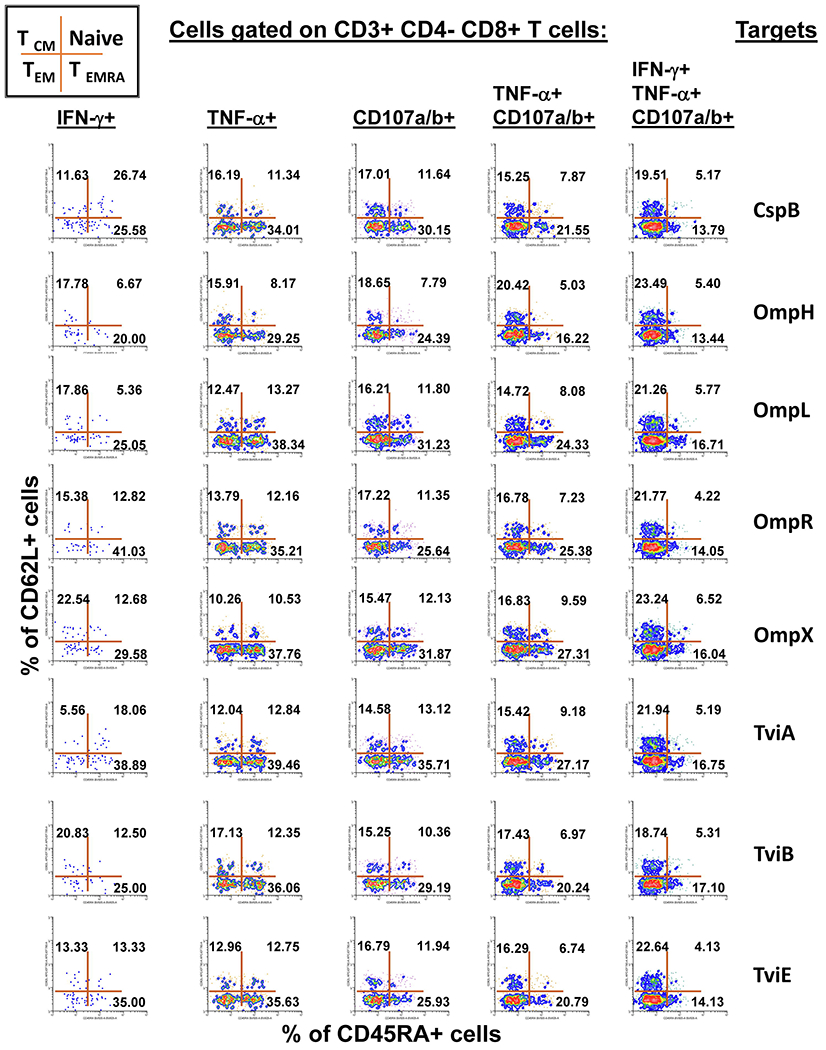
Subsequent gates on the mono and multifunctional CD8+ T-cell subsets described in Fig. 5 were used to identify the memory status among each of the 15 defined sub-phenotypes of CD8+ cells. The data is representative of one experiment/one volunteer showing mono-functional or multi-functional CD8+ T-cell responses to eight recombinant E. coli expressing S. Typhi/Hly (CspB, OmpH, OmpL, OmpR, OmpX, TviA, TviB and TviE) proteins at a multiplicity of infection (MOI) of 1:30. This figure illustrates a gating strategy in which CD8+ T-cells are further categorized based on the expression of CD45RA and CD62L markers. Cells in each resulting quadrant of the dot plot are then categorized in 4 subpopulations: central memory (CD45RA−CD62L+, TCM), naive (CD45RA+CD62L+, Naive), effector memory (CD45RA−CD62L−, TEM), effector memory expressing CD45RA (CD45RA+CD62L−, TEMRA). The 5 selected populations are those that were dominant in the volunteers who responded to stimulation with S. Typhi proteins, exhibiting sufficient number of positive events to allow downstream analyses. Numbers represent the percentage of positive cells in the respective quadrant.
Discussion
We have previously demonstrated the feasibility of using a recombinant E. coli expression system to uncover the antigen specificity of CD4+ and CD8+ T cells. Here, we expanded these studies to include the evaluation of 12 additional S. Typhi proteins: 4 outer membrane proteins (OmpH, OmpL, OmpR, OmpX), 3 Vi-polysaccharide biosynthesis proteins (TviA, TviB, TviE), 3 cold shock proteins (CspA, CspB, CspC), and 2 conserved hypothetical proteins (Chp 1 and 2), all selected based on the bioinformatic analyses of their putative T-cell epitope content. Although volunteers differed in their T-cell antigen specificity, T-cell immune responses against all 12 S. Typhi proteins were identified. Nine of these proteins, OmpH, OmpR, TviA, TviE, CspA, CspB, CspC, Chp 1 and Chp 2, have not been previously reported to trigger S. Typhi specific T-cell responses. These studies demonstrate the feasibility of using immunoinformatics analysis and the E. coli expressing system described here to uncover novel immunogenic T-cell proteins that could serve as targets for the production of protein-based vaccines. It is important to note that most, if not all, of the proteins selected in this manuscript play a role in S. Typhi survival, and therefore can be explored as promising vaccine antigens. For example, Omps represent a sophisticated macromolecular assembly that interact with a variety of host tissues for adhesion to and invasion of the cells [40]. Omps are also involved in the exchange of nutrients over the outer membrane of Gram-negative bacteria [40]. Indeed, porins such as OmpC, OmpF [41] and OmpS [42], are known to be potent immunogens with adjuvant properties. It is also worth mentioning that OmpX and OmpS of Salmonella Typhimurium have been shown to trigger T-cell immune responses in patients with Salmonella-induced reactive arthritis [43]. Interestingly, ompR positively regulates S. Typhi ompS2 porin gene [44]. The ompR gene also positively regulates the transcription of tviA and tviB genes [45] acting as an activator of Vi antigen [46]. TviA is also implicated in the reduction of IL-8 production by intestinal epithelial cells by repressing flagellin secretion [47]. Moreover, S. Typhi colonization provokes extensive transcriptional changes in genes related to Vi antigen biosyntheses (i.e., tviE), which appears to play a role in evasion of the mucosal immune defense [48, 49]. Finally, Csp family members are at high cellular abundance [50] with remarkable sequence conservation [51] suggesting essential functions in the bacteria. CspC appears to be crucial for stress resistance, motility, and biofilm formation [52].
Our study also confirms and expands our previous observation that upon stimulation by S. Typhi antigens the host mounts a coordinated, simultaneous and complementary response comprised of CD4+ and CD8+ cell responses to better fight the pathogen [33, 53]. While single cytokine producers characterized the majority of CD4+ T-cell responses, the frequency of cells expressing triple functions was higher among CD8+ T-cells. Of note, regardless of the T-cell subset studied, i.e., CD4+ or CD8+ T-cells, the concomitant expression of IFN-γ and IL-17A were at a very low levels or absent. These observation supplements previous data by Harrington et at. [54] and Park et at. [55] demonstrating that IFN-γ negatively regulates CD4+ T-cells production of IL-17 during the effector phase of the immune response. Interestingly, the profile of these responses by T-cell subsets depended only marginally, if at all, on the proteins evaluated in this study. We speculated that the quality and characteristics of the responses are largely determined by the T-cell subset, CD4+, and CD8+ T-cells, rather than the particular S. Typhi protein. These results also support previous data in a murine model showing that the depletion of either CD4+ or CD8+ T-cells had impaired recall immunity to oral challenge with the virulent S. Typhimurium at different times after vaccination [56].
One of the hallmarks of successful vaccination is the induction of strong and persistent memory T-cell responses [1]. In this study, we further characterized the cytokine production / CD107ab expression patterns of TCM, naïve, TEM, and TEMRA among CD4+ or CD8+ T-cells. We found that CD4+ or CD8+ T-cells are heterogeneous in terms of their memory phenotype, further suggesting the induction of complementary functions by these two major T-cell subsets. Consistent with the early descriptions of memory T-cell pool functions by Lanzavecchia, Sallusto and colleagues, CD8+ T-cells in our manuscript were mainly TEM and exhibited cytotoxic potential [57]. On the other hand, CD4+ T-cells with naïve and TEMRA phenotypes emerged as the most prominent producers of IL-17A as compared CD8+ T-cells. This profile demonstrates the plasticity of CD4+ T-cells and supports the view advanced by some studies showing that CD4+ T-cells producing IL-17 might represent a population of T-cells with stem cell-like properties capable of redirecting their functional programs, [58, 59]. Alternatively or concomitantly, certain culture conditions might be more prone to promote IL-17A secretion by naive CD4+ T-cells [60]. Thus, our results support the notion of the induction of differential functional characteristics among memory subsets, especially in terms of their capacity to produce cytokines and express cytotoxic markers (i.e., CD107 a/b). They also shed light on the relationship between the CD4+ or CD8+ T-cell subsets. However, this is a cross-sectional study following vaccination and therefore does not directly address the lineage relationships of TCM, naive, TEm, and TEMRA and their relative contributions in controlling S. Typhi infections. Moreover, these studies were performed with cells obtained at two-time points (before and 42 days after immunization). The characteristics of the responses by CD4+ and CD8+ T-cells in circulation might be different at other time points. Further studies in which the T-cell responses to the novel S. Typhi proteins reported in this study can be correlated with protection in human challenge studies and in which additional time points are examined will establish the significance of the current observations.
S. Typhi infects several types of cells including epithelial cells, macrophages, T- and B-cells [5, 6, 61–64]. Since the studies in this manuscript were performed exclusively with B-cells as target cells, it is important to note that peptide selection against an unique type of target cells may not represent the full spectrum of peptides likely to be presented by different cell types in each individual. As we observed for CD4+ and CD8+ cells, some peptides might be presented by many different cell types, while others migth be restricted to particular cells [65]. Antigen presentation is multifactorial, with highly diverse HLA-haplotypes among individuals, and cues from the microenvironment (e.g., systemic vs. mucosa, inflammatory vs. anti-inflammatory, and cross-talk between different cell types), all likely to affect how the host process and presents S. Typhi antigens [66]. Our data also suggest that vaccines directed to multiple proteins/antigens might be more immunogenic than those based on a single protein/antigen. This is perhaps not surprising since a single type of protein-derived peptide may not be present at levels sufficient, or exhibit the appropriate characteristics, to mount a protective response. Thus, the development of multi-component vaccines including many antigenic determinants, into vaccine formulations are more likely to succeed [27].
In conclusion, our study demonstrates a dichotomy between the functional characteristics of CD4+ or CD8+ T-cell subsets, suggestive of complementarity in terms of their memory differentiation, production of cytokines and cytotoxic capability. By defining phenotypic signatures for CD4+ or CD8+ memory subsets, we provide a framework for further characterization of S. Typhi-specific memory T cells to novel S. Typhi proteins. Those results also demonstrate the feasibility of using our antigen discovery platform, which might lead to the discovery of novel candidate vaccine antigens for S. Typhi, and perhaps other infectious organisms.
Supplementary Material
Supplementary Fig. 1. Magnitude of CD4+ and CD8+ T cell responses to targets infected with recombinant E. coli. Ex vivo PBMC from 15 volunteers collected before and 42 days after immunization were co-cultured for 16-18 hrs with autologous B-LCL targets infected at a 1:30 MOI with recombinant E. coli expressing Hly only or co-expressing S. Typhi antigens: CspA, CspB, OmpH, OmpL, OmpR, OmpX, TviA, TviB, TviE, and two conserved hypothetical proteins (Chp 1 and Chp2). After incubation, cells were stained, and the ability of the PBMC to produce one or more cytokines (IL-17A, IFN-γ and TNF-α) and/or express CD107a/b molecules was analyzed by flow cytometry. Two T-cell subset responses (i.e., CD4+ and CD8+ T cells) were evaluated. (A) Net responses were calculated by subtracting the T-cell responses to B-LCLs infected with recombinant E. coli expressing S. Typhi/Hly antigens from the responses of the controls (B-LCL expressing Hly only). Increases over day 0 were calculated by subtracting the net responses of the PBMC collected before immunization from the net responses of PBMC collected 42 days after immunization. (B) Correlation between CD4+ and CD8+ T cell responses. The data represent the combined results of 15 volunteers to all 12 proteins. Coefficients of correlation “R” and “P” values are shown. p values of <0.05 were considered statistically significant. Dashed lines represent 95% confidence intervals.
5. Acknowledgments
We are indebted to the volunteers who allowed us to perform this study. We also thank Mrs. Robin Barnes and the staff from the Recruiting Section of the Center for Vaccine Development for their help in collecting blood specimens and Dr. Haiyan Chen, and Mrs. Regina Harley and Catherine Storrer for excellent technical assistance.
This work was supported, in part, by National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Department of Health and Human Services (DHHS) federal research grants (https://www.niaid.nih.gov) R01 AI036525, and U19 AI082655 (Cooperative Center for Human Immunology [CCHI]) to M.B. Sztein; and U19 AI082642 (CCHI) to De Groot. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, the National Institutes of Health. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
7. References
- [1].Salerno-Goncalves R, Sztein MB. Cell-mediated immunity and the challenges for vaccine development. Trends in microbiology. 2006;14:536–42. [DOI] [PubMed] [Google Scholar]
- [2].Lundin BS, Johansson C, Svennerholm AM. Oral immunization with a Salmonella enterica serovar typhi vaccine induces specific circulating mucosa-homing CD4(+) and CD8(+) T cells in humans. Infect Immun. 2002;70:5622–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sheikh A, Khanam F, Sayeed MA, Rahman T, Pacek M, Hu Y, et al. Interferon-gamma and proliferation responses to Salmonella enterica Serotype Typhi proteins in patients with S. Typhi Bacteremia in Dhaka, Bangladesh. PLoS neglected tropical diseases. 2011;5:e1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sztein MB. Cell-mediated immunity and antibody responses elicited by attenuated Salmonella enterica Serovar Typhi strains used as live oral vaccines in humans. Clin Infect Dis. 2007;45 Suppl 1:S15–9. [DOI] [PubMed] [Google Scholar]
- [5].Salerno-Goncalves R, Fernandez-Vina M, Lewinsohn DM, Sztein MB. Identification of a human HLA-E-restricted CD8+ T cell subset in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J Immunol. 2004;173:5852–62. [DOI] [PubMed] [Google Scholar]
- [6].Salerno-Goncalves R, Pasetti MF, Sztein MB. Characterization of CD8(+) Effector T Cell Responses in Volunteers Immunized with Salmonella enterica Serovar Typhi Strain Ty21a Typhoid Vaccine. J Immunol. 2002;169:2196–203. [DOI] [PubMed] [Google Scholar]
- [7].Salerno-Goncalves R, Wahid R, Sztein MB. Immunization of volunteers with Salmonella enterica serovar Typhi strain Ty21a elicits the oligoclonal expansion of CD8+ T cells with predominant Vbeta repertoires. Infect Immun. 2005;73:3521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Salerno-Goncalves R, Wahid R, Sztein MB. Ex Vivo kinetics of early and long-term multifunctional human leukocyte antigen E-specific CD8+ cells in volunteers immunized with the Ty21a typhoid vaccine. Clinical and vaccine immunology : CVI. 2010;17:1305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sztein MB, Tanner MK, Polotsky Y, Orenstein JM, Levine MM. Cytotoxic T lymphocytes after oral immunization with attenuated vaccine strains of Salmonella typhi in humans. J Immunol. 1995;155:3987–93. [PubMed] [Google Scholar]
- [10].Viret JF, Favre D, Wegmuller B, Herzog C, Que JU, Cryz SJ Jr., et al. Mucosal and systemic immune responses in humans after primary and booster immunizations with orally administered invasive and noninvasive live attenuated bacteria. Infect Immun. 1999;67:3680–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wahid R, Salerno-Goncalves R, Tacket CO, Levine MM, Sztein MB. Cell-mediated immune responses in humans after immunization with one or two doses of oral live attenuated typhoid vaccine CVD 909. Vaccine. 2007;25:1416–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wahid R, Salerno-Goncalves R, Tacket CO, Levine MM, Sztein MB. Generation of specific effector and memory T cells with gut- and secondary lymphoid tissue-homing potential by oral attenuated CVD 909 typhoid vaccine in humans. Mucosal Immunology. 2008;1:389–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].McArthur MA, Sztein MB. Heterogeneity of multifunctional IL-17A producing S. Typhi-specific CD8+ T cells in volunteers following Ty21a typhoid immunization. PLoS One. 2012;7:e38408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Eloe-Fadrosh EA, McArthur MA, Seekatz AM, Drabek EF, Rasko DA, Sztein MB, et al. Impact of oral typhoid vaccination on the human gut microbiota and correlations with s. Typhi-specific immunological responses. PLoS One. 2013;8:e62026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kirkpatrick BD, Tenney KM, Larsson CJ, O’Neill JP, Ventrone C, Bentley M, et al. The novel oral typhoid vaccine M01ZH09 is well tolerated and highly immunogenic in 2 vaccine presentations. J Infect Dis. 2005;192:360–6. [DOI] [PubMed] [Google Scholar]
- [16].Sztein MB. Is a Human CD8 T-Cell Vaccine Possible, and if So, What Would It Take? CD8 T-Cell-Mediated Protective Immunity and Vaccination against Enteric Bacteria. Cold Spring Harbor perspectives in biology. 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fresnay S, McArthur MA, Magder L, Darton TC, Jones C, Waddington CS, et al. Salmonella Typhi-specific multifunctional CD8+ T cells play a dominant role in protection from typhoid fever in humans. Journal of translational medicine. 2016;14:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fresnay S, McArthur MA, Magder LS, Darton TC, Jones C, Waddington CS, et al. Importance of Salmonella Typhi-Responsive CD8+ T Cell Immunity in a Human Typhoid Fever Challenge Model. Front Immunol. 2017;8:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bumann D Identification of Protective Antigens for Vaccination against Systemic Salmonellosis. Front Immunol. 2014;5:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Barat S, Willer Y, Rizos K, Claudi B, Maze A, Schemmer AK, et al. Immunity to intracellular Salmonella depends on surface-associated antigens. PLoS Pathog. 2012;8:e1002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Napolitani G, Kurupati P, Teng KWW, Gibani MM, Rei M, Aulicino A, et al. Clonal analysis of Salmonella-specific effector T cells reveals serovar-specific and cross-reactive T cell responses. Nat Immunol. 2018;19:742–54. [DOI] [PubMed] [Google Scholar]
- [22].Song J, Gao X, Galan JE. Structure and function of the Salmonella Typhi chimaeric A(2)B(5) typhoid toxin. Nature. 2013;499:350–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li W, Joshi MD, Singhania S, Ramsey KH, Murthy AK. Peptide Vaccine: Progress and Challenges. Vaccines. 2014;2:515–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sheikh A, Charles RC, Rollins SM, Harris JB, Bhuiyan MS, Khanam F, et al. Analysis of Salmonella enterica serotype paratyphi A gene expression in the blood of bacteremic patients in Bangladesh. PLoS neglected tropical diseases. 2010;4:e908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bouwer HG, Alberti-Segui C, Montfort MJ, Berkowitz ND, Higgins DE. Directed antigen delivery as a vaccine strategy for an intracellular bacterial pathogen. Proc Natl Acad Sci U S A. 2006;103:5102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hu PQ, Tuma-Warrino RJ, Bryan MA, Mitchell KG, Higgins DE, Watkins SC, et al. Escherichia coli expressing recombinant antigen and listeriolysin O stimulate class I-restricted CD8+ T cells following uptake by human APC. J Immunol. 2004;172:1595–601. [DOI] [PubMed] [Google Scholar]
- [27].Salerno-Goncalves R, Tettelin H, Lou D, Steiner S, Rezwanul T, Guo Q, et al. Use of a novel antigen expressing system to study the Salmonella enterica serovar Typhi protein recognition by T cells. PLoS neglected tropical diseases. 2017;11:e0005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Terry FE, Moise L, Martin RF, Torres M, Pilotte N, Williams SA, et al. Time for T? Immunoinformatics addresses vaccine design for neglected tropical and emerging infectious diseases. Expert review of vaccines. 2015;14:21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Moise L, Gutierrez A, Kibria F, Martin R, Tassone R, Liu R, et al. iVAX: An integrated toolkit for the selection and optimization of antigens and the design of epitope-driven vaccines. Human vaccines & immunotherapeutics. 2015;11:2312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH) adopts Consolidated Guideline on Good Clinical Practice in the Conduct of Clinical Trials on Medicinal Products for Human Use. International digest of health legislation. 1997;48:231–4. [PubMed] [Google Scholar]
- [31].Levine MM, Ferreccio C, Abrego P, Martin OS, Ortiz E, Cryz S. Duration of efficacy of Ty21a, attenuated Salmonella typhi live oral vaccine. Vaccine. 1999;17:S22–7. [DOI] [PubMed] [Google Scholar]
- [32].Levine MM, Ferreccio C, Black RE, Germanier R. Large-scale field trial of Ty21a live oral typhoid vaccine in enteric-coated capsule formulation. Lancet. 1987;1:1049–52. [DOI] [PubMed] [Google Scholar]
- [33].Salerno-Goncalves R, Wyant TL, Pasetti MF, Fernandez-Vina M, Tacket CO, Levine MM, et al. Concomitant Induction of CD4(+) and CD8(+) T Cell Responses in Volunteers Immunized with Salmonella enterica Serovar Typhi Strain CVD 908-htrA. J Immunol. 2003;170:2734–41. [DOI] [PubMed] [Google Scholar]
- [34].Baker S, Dougan G. The genome of Salmonella enterica serovar Typhi. Clin Infect Dis. 2007;45 Suppl 1:S29–33. [DOI] [PubMed] [Google Scholar]
- [35].Deng W, Liou SR, Plunkett G 3rd, Mayhew GF, Rose DJ, Burland V, et al. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J Bacteriol. 2003;185:2330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Coren LV, Shatzer T, Ott DE. CD45 immunoaffinity depletion of vesicles from Jurkat T cells demonstrates that exosomes contain CD45: no evidence for a distinct exosome/HIV-1 budding pathway. Retrovirology. 2008;5:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, et al. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods. 2003;281:65–78. [DOI] [PubMed] [Google Scholar]
- [38].Lamoreaux L, Roederer M, Koup R. Intracellular cytokine optimization and standard operating procedure. Nature protocols. 2006;1:1507–16. [DOI] [PubMed] [Google Scholar]
- [39].Booth JS, Toapanta FR, Salerno-Goncalves R, Patil S, Kader HA, Safta AM, et al. Characterization and functional properties of gastric tissue-resident memory T cells from children, adults, and the elderly. Front Immunol. 2014;5:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Galdiero S, Falanga A, Cantisani M, Tarallo R, Della Pepa ME, D’Oriano V, et al. Microbe-host interactions: structure and role of Gram-negative bacterial porins. Current protein & peptide science. 2012;13:843–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Perez-Toledo M, Valero-Pacheco N, Pastelin-Palacios R, Gil-Cruz C, Perez-Shibayama C, Moreno-Eutimio MA, et al. Salmonella Typhi Porins OmpC and OmpF Are Potent Adjuvants for T-Dependent and T-Independent Antigens. Front Immunol. 2017;8:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Moreno-Eutimio MA, Tenorio-Calvo A, Pastelin-Palacios R, Perez-Shibayama C, Gil-Cruz C, Lopez-Santiago R, et al. Salmonella Typhi OmpS1 and OmpS2 porins are potent protective immunogens with adjuvant properties. Immunology. 2013;139:459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Singh R, Shasany AK, Aggarwal A, Sinha S, Sisodia BS, Khanuja SP, et al. Low molecular weight proteins of outer membrane of Salmonella typhimurium are immunogenic in Salmonella induced reactive arthritis revealed by proteomics. Clin Exp Immunol. 2007;148:486–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fernandez-Mora M, Puente JL, Calva E. OmpR and LeuO positively regulate the Salmonella enterica serovar Typhi ompS2 porin gene. J Bacteriol. 2004;186:2909–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang Y, Xia L, Lin L, Tang H, Osei-Adjei G, Xu S, et al. Reciprocal Regulation of OmpR and Hfq and Their Regulatory Actions on the Vi Polysaccharide Capsular Antigen in Salmonella enterica Serovar Typhi. Current microbiology. 2018;75:773–8. [DOI] [PubMed] [Google Scholar]
- [46].Pickard D, Li J, Roberts M, Maskell D, Hone D, Levine M, et al. Characterization of defined ompR mutants of Salmonella typhi: ompR is involved in the regulation of Vi polysaccharide expression. Infect Immun. 1994;62:3984–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Winter SE, Raffatellu M, Wilson RP, Russmann H, Baumler AJ. The Salmonella enterica serotype Typhi regulator TviA reduces interleukin-8 production in intestinal epithelial cells by repressing flagellin secretion. Cell Microbiol. 2008;10:247–61. [DOI] [PubMed] [Google Scholar]
- [48].Nickerson KP, Senger S, Zhang Y, Lima R, Patel S, Ingano L, et al. Salmonella Typhi Colonization Provokes Extensive Transcriptional Changes Aimed at Evading Host Mucosal Immune Defense During Early Infection of Human Intestinal Tissue. EBioMedicine. 2018;31:92–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Galen JE, Buskirk AD, Tennant SM, Pasetti MF. Live Attenuated Human Salmonella Vaccine Candidates: Tracking the Pathogen in Natural Infection and Stimulation of Host Immunity. EcoSal Plus. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, et al. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics. 2008;9:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yamanaka K, Fang L, Inouye M. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol. 1998;27:247–55. [DOI] [PubMed] [Google Scholar]
- [52].Michaux C, Holmqvist E, Vasicek E, Sharan M, Barquist L, Westermann AJ, et al. RNA target profiles direct the discovery of virulence functions for the cold-shock proteins CspC and CspE. Proc Natl Acad Sci U S A. 2017;114:6824–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Booth JS, Goldberg E, Patil SA, Greenwald BD, Sztein MB. Association between S. Typhi-specific memory CD4+ and CD8+ T responses in the terminal ileum mucosa and in peripheral blood elicited by the live oral typhoid vaccine Ty21a in humans. Human vaccines & immunotherapeutics. 2019:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–32. [DOI] [PubMed] [Google Scholar]
- [55].Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mastroeni P, Villarreal-Ramos B, Hormaeche CE. Role of T cells, TNF alpha and IFN gamma in recall of immunity to oral challenge with virulent salmonellae in mice vaccinated with live attenuated aro- Salmonella vaccines. Microbial pathogenesis. 1992;13:477–91. [DOI] [PubMed] [Google Scholar]
- [57].Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–63. [DOI] [PubMed] [Google Scholar]
- [58].Muranski P, Borman ZA, Kerkar SP, Klebanoff CA, Ji Y, Sanchez-Perez L, et al. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity. 2011;35:972–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–55. [DOI] [PubMed] [Google Scholar]
- [60].Muranski P, Restifo NP. Essentials of Th17 cell commitment and plasticity. Blood. 2013;121:2402–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Salerno-Goncalves R, Rezwan T, Sztein MB. B cells modulate mucosal associated invariant T cell immune responses. Front Immunol. 2014;4:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Salerno-Goncalves R, Kayastha D, Fasano A, Levine MM, Sztein MB. Crosstalk between leukocytes triggers differential immune responses against Salmonella enterica serovars Typhi and Paratyphi. PLoS neglected tropical diseases. 2019;13:e0007650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Salerno-Gongalves R, Galen JE, Levine MM, Fasano A, Sztein MB. Manipulation of Salmonella Typhi Gene Expression Impacts Innate Cell Responses in the Human Intestinal Mucosa. Frontiers in Immunology. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Salerno-Goncalves R, Fasano A, Sztein MB. Engineering of a multicellular organotypic model of the human intestinal mucosa. Gastroenterology. 2011;141:e18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Takahashi S, Mok H, Parrott MB, Marini FC 3rd, Andreeff M, Brenner MK, et al. Selection of chronic lymphocytic leukemia binding peptides. Cancer Res. 2003;63:5213–7. [PubMed] [Google Scholar]
- [66].McConnell SC, Hernandez KM, Wcisel DJ, Kettleborough RN, Stemple DL, Yoder JA, et al. Alternative haplotypes of antigen processing genes in zebrafish diverged early in vertebrate evolution. Proc Natl Acad Sci U S A. 2016;113:E5014–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Magnitude of CD4+ and CD8+ T cell responses to targets infected with recombinant E. coli. Ex vivo PBMC from 15 volunteers collected before and 42 days after immunization were co-cultured for 16-18 hrs with autologous B-LCL targets infected at a 1:30 MOI with recombinant E. coli expressing Hly only or co-expressing S. Typhi antigens: CspA, CspB, OmpH, OmpL, OmpR, OmpX, TviA, TviB, TviE, and two conserved hypothetical proteins (Chp 1 and Chp2). After incubation, cells were stained, and the ability of the PBMC to produce one or more cytokines (IL-17A, IFN-γ and TNF-α) and/or express CD107a/b molecules was analyzed by flow cytometry. Two T-cell subset responses (i.e., CD4+ and CD8+ T cells) were evaluated. (A) Net responses were calculated by subtracting the T-cell responses to B-LCLs infected with recombinant E. coli expressing S. Typhi/Hly antigens from the responses of the controls (B-LCL expressing Hly only). Increases over day 0 were calculated by subtracting the net responses of the PBMC collected before immunization from the net responses of PBMC collected 42 days after immunization. (B) Correlation between CD4+ and CD8+ T cell responses. The data represent the combined results of 15 volunteers to all 12 proteins. Coefficients of correlation “R” and “P” values are shown. p values of <0.05 were considered statistically significant. Dashed lines represent 95% confidence intervals.


