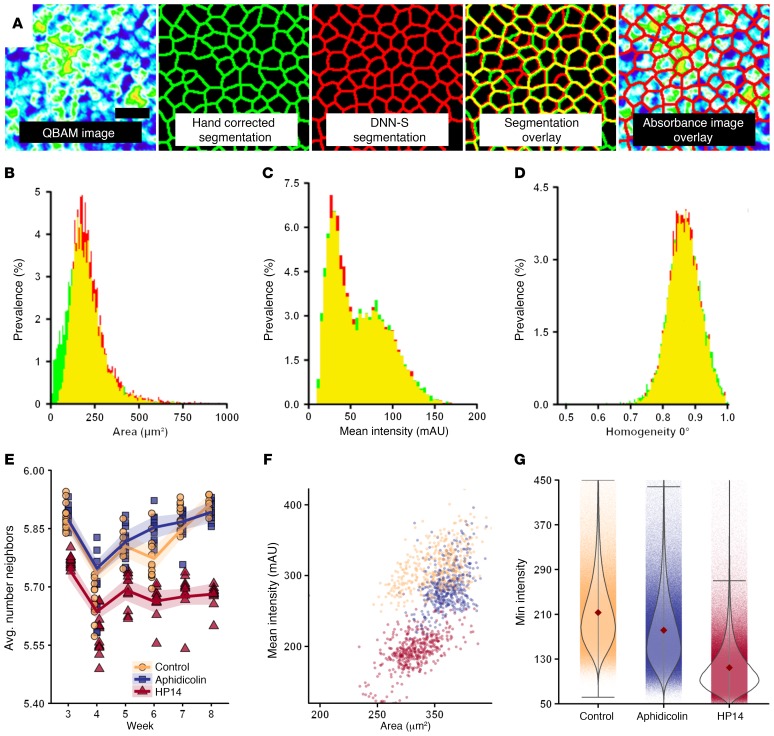
Figure 4. DNN segmentation of iPSC-RPE in QBAM images.
(A) A DNN (DNN-S) was constructed to segment iPSC-RPE cells in absorbance images. To train the DNN, iPSC-RPE monolayers were fluorescently labeled for cell borders (ZO-1) and registered to absorbance images for hand labeling of cell borders. Scale bar: 25 μm. (B–D) A comparison of 3 of 42 cell feature histograms for hand segmented (green) and DNN segmented (red) images, where yellow is the overlap in the histograms. (E) Time course of the average number of cells bordering each cell for an entire well. Shaded region represents 95% SEM. Twelve wells per time per treatment are shown. (F) A scatterplot of mean cell area and mean intensity (absorbance) for each treatment group assessed for each microscope FOV. Each dot represents 1 of 864 fields of view (12 wells per treatment, 12 images per well, 6 time points). (G) Minimum intensity (absorbance) found for individual cells as a function of treatment. Whiskers represent 3 times SD and single dots behind the violin represent individual cells measured. n = 3,871,106 cells for control; n = 3,831,362 cells for aphidicolin; n = 4,146,927 cells for HPI4. A complete set of feature histogram comparisons is presented in Supplemental Figure 5, and results of statistical tests are in Supplemental Table 2. Red diamond indicates the mean.