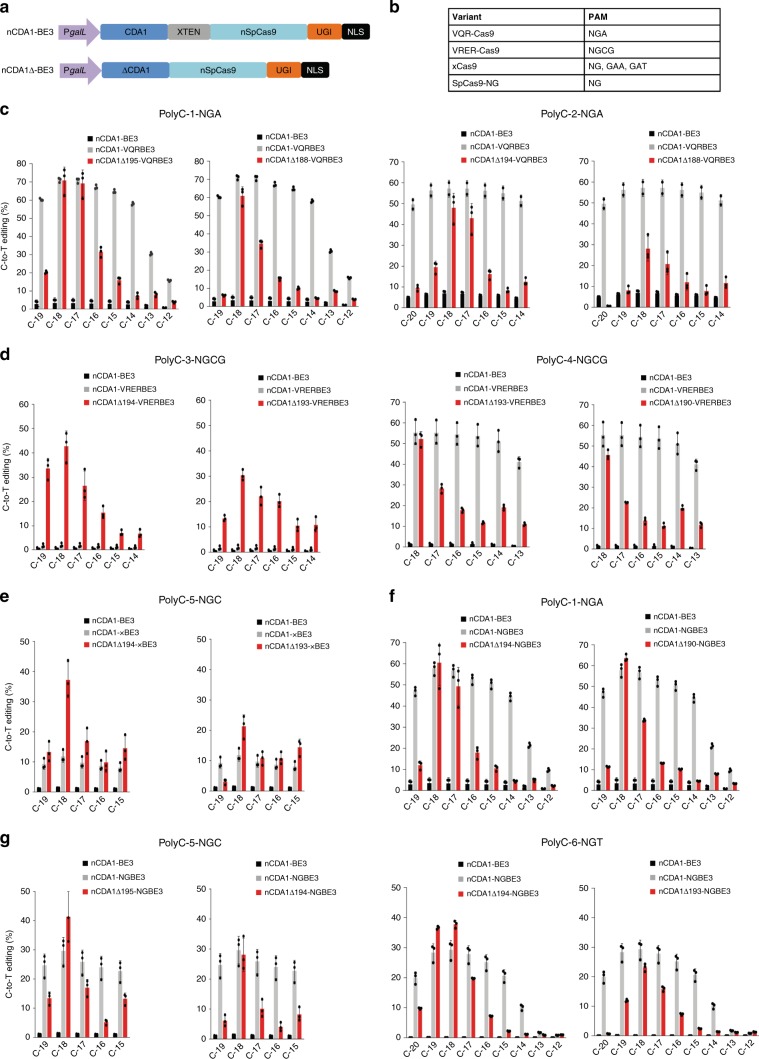
Fig. 1. High-precision base editing at target sites containing non-NGG PAMs.
a Structure of nCDA1-BE3 in comparison to base editors harboring CDA1 truncations (ΔCDA1). nSpCas9: Streptococcus pyogenes Cas9 nickase; XTEN: synthetic linker sequence;15 UGI: uracil DNA glycosylase inhibitor; NLS: nuclear localization signal. b Cas9 variants with altered PAM specificities. c–g BE variants with CDA1 truncations mediate high-precision base editing at target sites comprised of multiple cytidines (polyC targets; for sequences see Supplementary Table 1). The x-axis shows the Cs in the target sequence with their position relative to the PAM indicated (Supplementary Table 1). The y-axis (C-to-T editing in %) represents the percentage of total sequencing reads with the target C converted to T. c Analysis of base editing precision of VQR-Cas9 BEs fused to selected C-terminally truncated versions of CDA1 (for the complete deletion series, see Supplementary Fig. 1). For comparison, the BE carrying the full-length CDA1 and the nCDA1-BE3 editor are also included. d Analysis of base editing precision of VRER-Cas9 BEs fused to C-terminally truncated CDA1 versions (for the complete deletion series, see Supplementary Fig. 2). e Analysis of base editing precision of xCas9 BEs fused to C-terminally truncated CDA1 versions (for the complete deletion series, see Supplementary Fig. 3). f, g Analysis of base editing precision of SpCas9-NG BEs fused to C-terminally truncated CDA1 versions (for the complete deletion series, see Supplementary Fig. 4). Values and error bars represent the mean and standard deviation of three independent biological replicates. Source data underlying c–g are provided as a Source Data file.