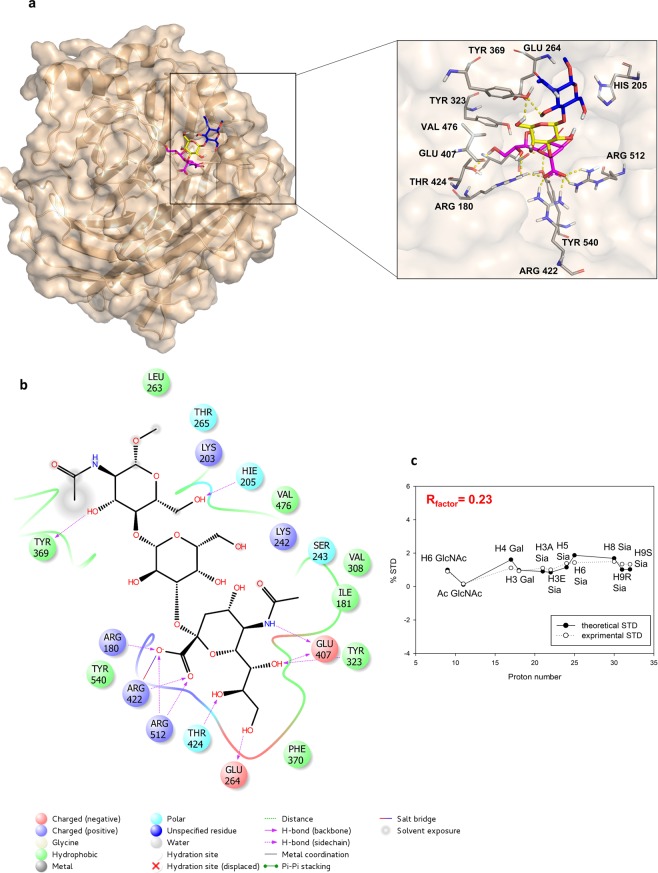
Figure 5.
3D model of MuV-HN in complex with trisaccharide 1 and CORCEMA analysis. (a) Close up view of ligand 1 binding mode at the MuV-HN active site, drawn by using Pymol 2.3 Software (https://pymol.org/2/). The main amino acid residues involved in the binding are represented in stick. Galactose and N-acetyl Glucosamine residues are depicted in yellow and blue, respectively. (b) Two-dimensional plot illustrating the interactions of the sialylated trisaccharide 1 with the residues in the binding pocket of MuV-HN derived by a tool included in Maestro Version 10.5.014 (https://www.schrodinger.com/maestro). Dotted arrows represent hydrogen bonds with functional groups from side chains and solid arrows such with functional groups of the backbone. The residues shown, close to the ligand, are involved into hydrophobic and polar interactions. (c) Comparison between experimental (dashed line) and theoretical (solid line) STD data for the selected 3D complex of the interaction between trisaccharide 1 and MuV-HN head domain.