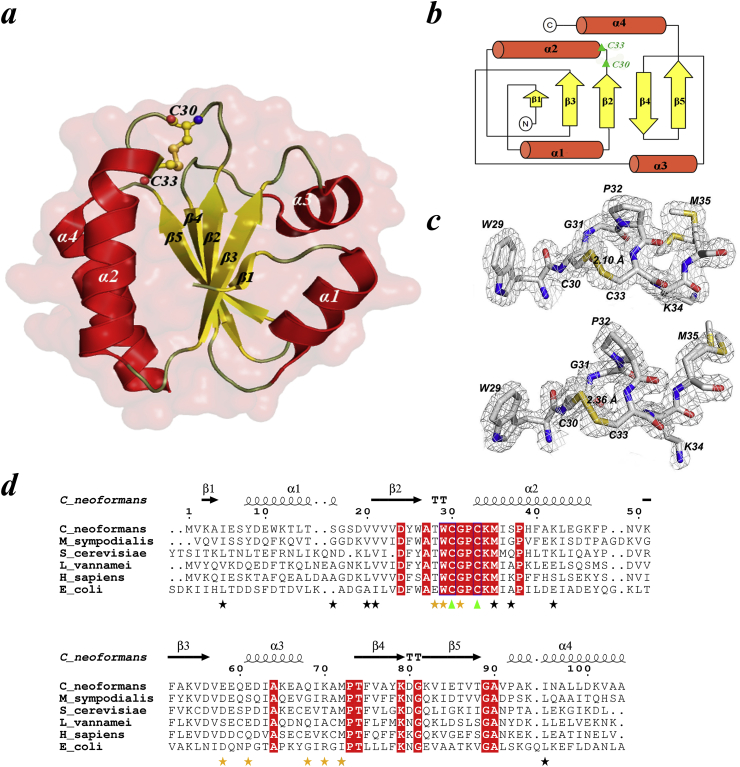
Fig. 1.
General architecture of CnTrx1. (a) Ribbon diagram of the overall structure of CnTrx1 with secondary structure elements labelled. Catalytic cysteines Cys30 and Cys33 are shown in the oxidized conformation observed in the crystal. All structure figures were made using PyMOL [55]. (b) Topological diagram of the CnTrx1 fold, showing the five-stranded mixed beta sheet sandwiched by four alpha helices. (c) Electron density 2|Fo| –|Fc| map, contoured at 1.0 σ around the conserved active site region WCGPCKM motif of CnTrx1 chain A (top) and B (bottom). The continuous electron density presents the disulfide bond between Sγ atoms of Cys30 and Cys33. (d) Sequence alignment of CnTrx1 (5JY5) with sequences of other thioredoxin proteins of known structure from M. sympodialis (2J23), S. cerevisiae (2OE0), L. vannamei (3ZZX), H. Sapiens(5DQY) and E. coli (1XOA). Absolutely conserved active-site cysteines are indicated with green triangle. Identical residues are in red boxes. Secondary structure elements of the CnTrx1 are shown above the alignment. The yellow stars bellow the residues indicate side chains involve hydrogen bonds between monomers, while black stars indicate those that have double conformation (Thr28A and Met72A,B also have double conformation). The alignment was performed using CLUSTAL W [56] and the figure was prepared with ESPript [57]. His-tag was removed from the sequence.