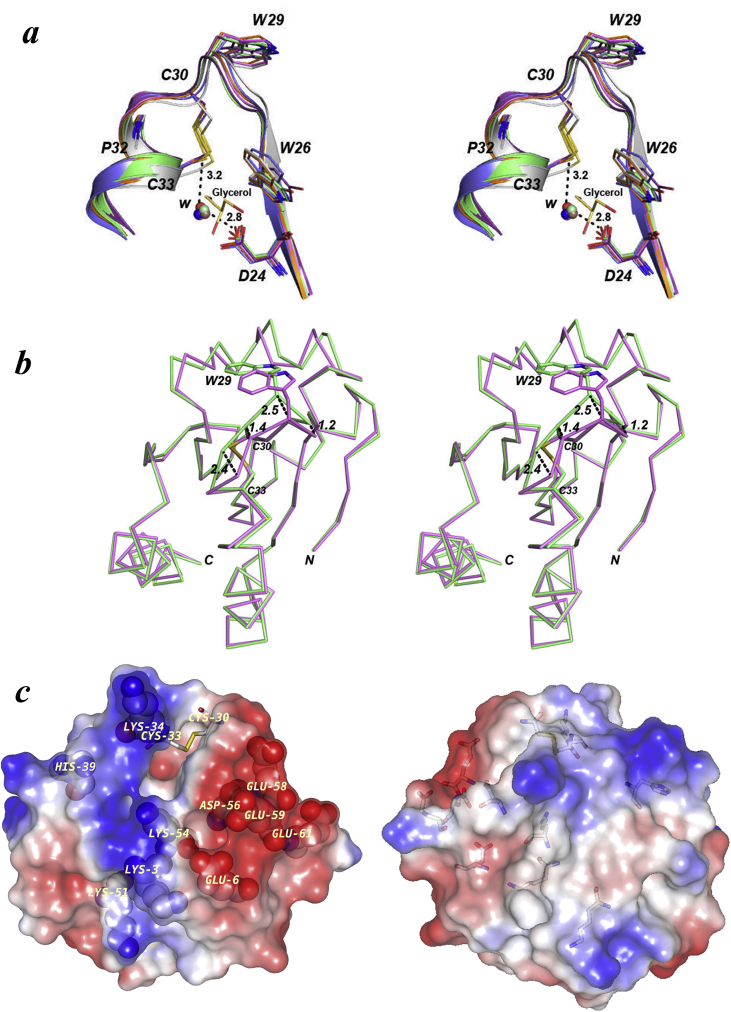
Fig. 3.
Superposition of the active site motif from different thioredoxins and Stereo surface representation of CnTrx1 monomer. (a) Trxs C. neoformans monomer B (orange, PDB ID: 5JY5), M. sympodialys (blue, PDB ID: 2J23), S. cerevisiae (purple, PDB ID: 2OE0), L. vannamei (green, PDB ID: 4AJ6), H. Sapiens (pink, PDB ID: 5DQY) and E. coli (gray, PDB ID: 1XOA). The overlay shows the conservation around the cysteines and the position of water molecules bound to Asp. The distances indicated between the internal water molecule with Asp24 and Cys33 are for the CnTrx1. (b) Superposition of CnTrx1ox monomers A (green) and B (pink) with the side chains of Trp29, Cys30 and Cys33 side chains shown in stick. The distances between Cα atoms of the two monomers in the loop region prior to helix α2 are depicted by black dashes and given in Å. (c) Two 180° apart views of the CnTrx1 electrostatic surface potentials (red, negative; blue, positive) as calculated with the GRASP Poisson-Boltzmann [58]. The disulfide bond (Cys30-Cys33) surface was omitted to facilitate viewing. The charged amino acid residues are pointed out by their type and sequence number. The figures were made using PyMOL.