Fig. 1.
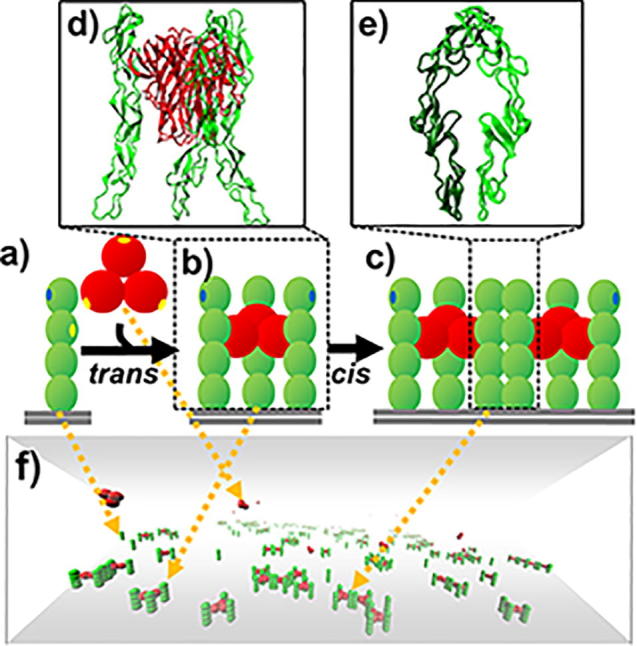
A coarse-grained model was constructed to simulate the spatial-temporal process of binding between TNF ligands and their receptors. (a) The trimeric TNF ligand is represented by three rigid bodies that are spatially arranged into a three-fold symmetry, while CRD domains in a TNF receptor are spatially aligned into a rod-like shape. (b) Through the trans-binding sites (yellow dots) on the surface of each ligand subunit and the second N-terminal domain of receptor, a ligand can simultaneously bind to three receptors and form a basic unit of signaling complex. (c) Furthermore, cis-binding site (blue dots) is assigned on the surface of the N-terminal domains in TNF receptors so they can be laterally connected together. (d) The x-ray crystal structures of ligands and receptors that form trans-interactions. (e) The x-ray crystal structures of two receptors that form cis-interactions. (f) Finally, a kinetic Monte-Carlo algorithm is applied to simulate the system, in which TNF receptors are distributed on the plasma membrane represented by the bottom surface of a three-dimensional simulation box. The space above the plasma membrane represents the extracellular region, where TNF ligands are located. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
