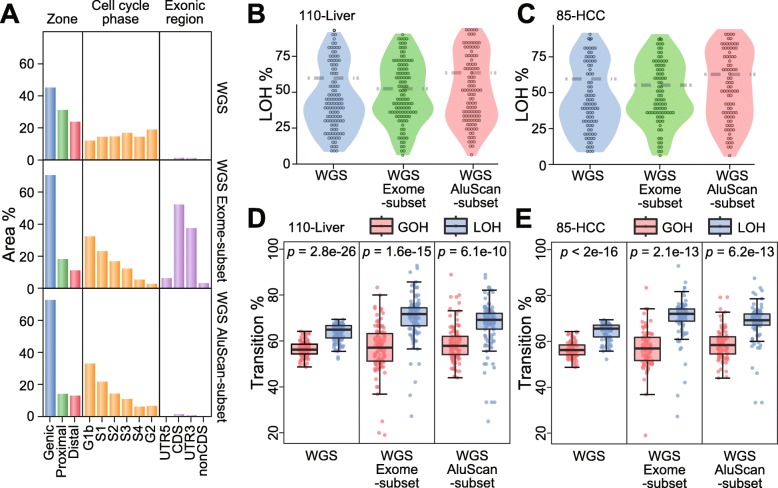
Fig. 2.
Comparison of the properties of WGS sequences of the 110-Liver cohort with the Exome-subset and the AluScan-subset. (a) Distribution of WGS, Exome-subset and AluScan sequences (in % of sequence regions) among the Genic, Proximal and Distal zones, with p values denoting the difference between GOHs and LOHs. among different cell cycle phases; and among different exonic regions. (b, c) Violin plots of LOH% (percentages of somatic LOHs relative to SNV load) in the 110-Liver and 85- HCC cohorts for three types of sequence data. (d, e) The percentages of transitional somatic GOHs and LOHs relative to total somatic GOHs and LOHs (red and blue boxes) for three types of sequence data, with the significant difference between GOHs and LOHs from paired t-tests expressed by the p-values for the 110-Liver and 85-HCC cohorts