Graphical abstract
Method name: Generation of SH-SY5Y-CRE reporter cell line
Keywords: Viral vectors, Fluorescence, Transduction
Abstract
Adenovirus-associated virus is a powerful vector system for transducing cells in vivo. It is widely used in animal systems due to high transduction efficiency of non-dividing cells with more than a dozen serotypes that have preferential tissue tropism. The viral genome remains episomal in the nucleus but maintains sustained expression in terminally differentiated cells for several weeks to months. Despite the popularity of recombinant AAV (rAAV) vectors, quality control testing of the virus after production is largely limited to physical characteristics such as viral genomes/ml determinations and silver staining acrylamide gels to determine purity. Functional testing, in vivo, is not practical due to high cost and restricted access of animal care and long duration of the assay (2–3 weeks). Some functional testing can be accomplished in cultured cells such as HEK293 cells, but HEK293 cells limit the types of rAAV constructs that can be tested. Many rAAV constructs are designed to study neurons in the brain with neural-specific promoters and many are floxed with loxp sites to be “activated” only in Cre-expressing neurons in transgenic animals. To develop a reporter cell line for rapid rAAV quality control assessment of these neural-specific, floxed rAAV constructs, we used the lentiviral system to stably express Cre recombinase in the SH-SY5Y neuroblastoma cell line.
-
•
A simple and economic method to evaluate recombinant AAV in vitro.
-
•
Allows functional validation of rAAV across a wide range of serotypes and promoters.
-
•
Allows functional validation of Cre-dependent rAAV constructs.
Specification Table
| Subject Area: | Biochemistry, Genetics and Molecular Biology |
| More specific subject area: | Recombinant Viral Vectors |
| Method name: | Generation of SH-SY5Y-CRE reporter cell line. |
| Name and reference of original method: | N/A |
| Resource availability: | N/A |
Methods details
Background
AAV vectors are often the system of choice when transducing cells in vivo. However, rAAV transduces different cell lines in culture to varying efficiencies which complicates functional validation of rAAV in cell lines [1]. Furthermore, different rAAV serotypes have cell preferences which transduce cells at different efficiencies. Although, HEK293 cells do work well across multiple serotypes with good efficiencies, it is an inadequate cell line for testing neuronal specific constructs or floxed constructs that are only expressed upon Cre recombination.
Generation of SH-SY5Y-CRE expressing cells
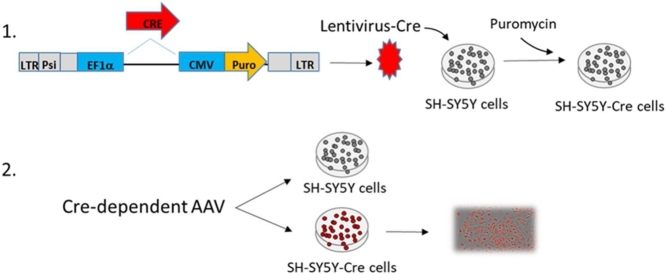
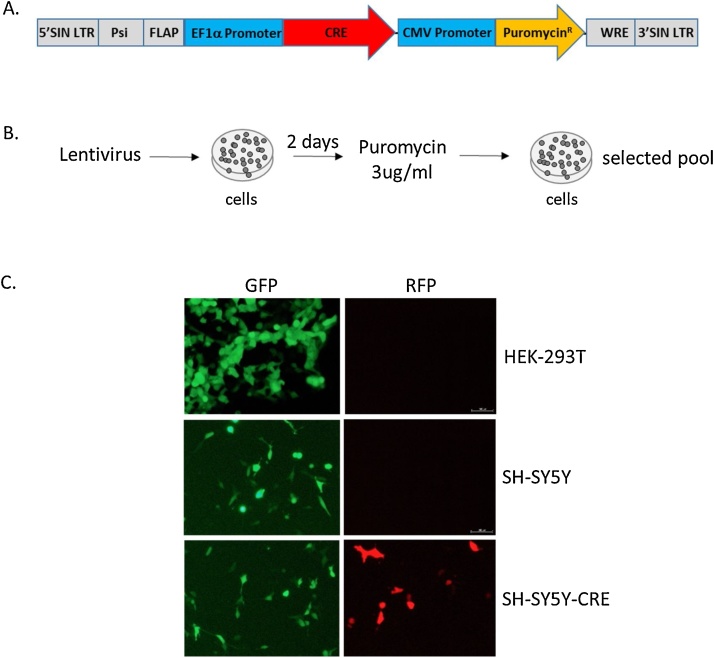
To develop a reporter cell line capable of rapidly and economically validating neuronal specific, floxed rAAV vectors, we used the lentiviral system to stably express Cre in the neuroblastoma SH-SY5Y cell line [2,3]. To generate the lentivirus, we inserted a codon optimized Cre recombinase gene into the SphI and NotI sites of the pLentilox EF1α-Puro vector (University of Michigan Vector Core, plasmid and sequence map are available upon request) generating a construct with EF1α promoter-driven Cre expression and CMV promoter-driven puromycin resistance (pLentilox EF1α-Cre-CMV-Puro, plasmid and sequence map are available upon request; Fig. 1A). Similar constructs can be found on Addgene.org. The EF1α promoter was chosen due to its consistently strong expression across a wide range of cell types and resistance to being silenced over time by epigenetic factors such as DNA methylation [4,5]. The construct was then packaged into lentivirus. Lentiviral packaging vectors psPAX2 (1.25μg), and pC1-VSVG (1.25μg) with pLentilox EF1α-Cre-CMV-Puro (2.5μg) transfer plasmid were incubated with PEI (15μg; molecular weight 25000, Polysciences, Inc) in 0.5mls of Optimem (Life Technologies) at room temperature for 20 min. Three milliliters of complete DMEM [Gibco, Cat #11965; 10% FBS (Hyclone) and 1x GlutaMAX (Gibco)] was added to the transfection mix and was added to a T25 tissue culture flask (Falcon) of 80% confluent HEK293T cells. Supernatant was collected after 72 h and cell debris was pelleted by centrifugation at 2000 rpms (805xg rcf) in an Eppendorf 5810R table top centrifuge at 4 °C for 10 min. The lentiviral supernatant was stored in 1ml aliquots at −80 °C. Virus transductions were performed on SH-SY5Y cells (Cat. # CRL-2266) obtained from American Type Culture Collection (ATCC), which were seeded on 6 well plates (∼2.5 × 105 cells/well) one day prior to lentiviral transduction (Fig. 1B). Cell media was changed to 1.5ml of viral supernatant (3 MOI) containing 8μg/ml Polybrene (Sigma). The cells were then incubated at 37 °C with 5% CO2 for 48 h prior to selection with 3μg/ml puromycin (Fisher Scientific) in complete media (DMEM with 10 % FBS, 1x non-essential amino acids and 1x sodium pyruvate) for 4 days. SH-SY5Y parental cells were all dead after 4 days of puromycin selection. Surviving SH-SY5Y-CRE cells were then expanded and stored in liquid nitrogen.
Fig. 1.
Generation of the SH-SY5Y-CRE reporter cell line. A. A schematic representation of the lentiviral construct used to stably express CRE in SH-SY5Y cells. B. SH-SY5Y cells were transduced with Lenti-EF1α-CRE-CMV-Puro-VSVG and selected with 3μg/ml puromycin for 4 days. C. The puromycin-resistant cells were transfected with a plasmid containing a floxed eGFP cassette followed by an mCherry cDNA. In the presence of active CRE, the eGFP cassette is removed and mCherry expression is activated. Expression of mCherry is only observed in SH-SY5Y-CRE cells (bottom panels) and not in SH-SY5Y parental cells (middle panels) or HEK293T cells (top panels). All images were taken 3 days post transfection at 20x magnification on an Olympus IX7 UV inverted microscope.
Functional Cre expression in SH-SY5Y-CRE cells
Puromycin resistant SH-SY5Y-CRE cells were transfected with a Cre-reporter plasmid (pCAG-fGFPf-RFP, gift from Kenneth Kwan, University of Michigan). This reporter plasmid has a CAG promoter followed by a floxed GFP cDNA and mCherry cDNA. The GFP cDNA prevents expression of mCherry unless the GFP is removed by Cre-recombination. HEK293T, SH-SY5Y and SH-SY5Y-CRE cells were transfected with just the Cre-reporter plasmid (0.3μg DNA with 1ug PEI in 0.3 ml of Optimem per well). Three days post-transfection, only GFP can be detected in the HEK293T or SH-SY5Y cells (Fig. 1C). However, in the SH-SY5Y-CRE cells mCherry can be detected suggesting Cre recombinase is expressed and functional (Fig. 1C). For quantitative analyses of CRE-dependent rAAVs, this method can be adapted by clonally isolating SH-SY5Y-CRE cells and screening for high CRE activity.
Characterization of SH-SY5Y-CRE as an rAAV reporter cell line
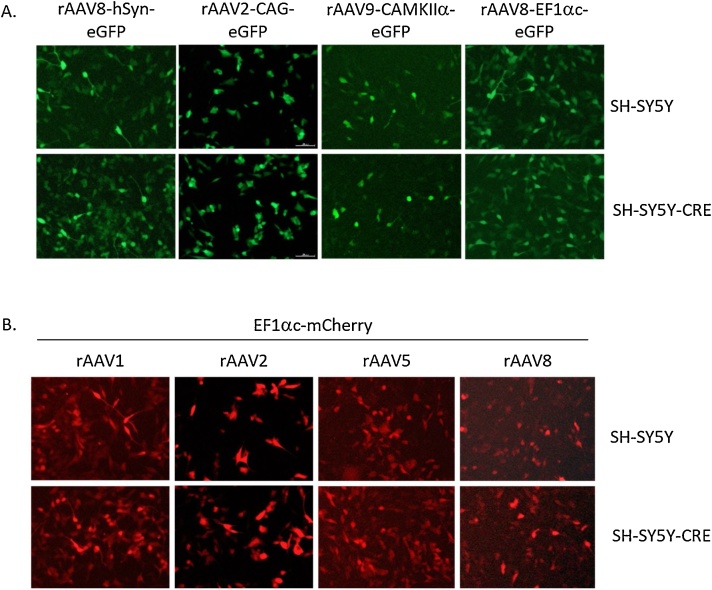
To characterize the SH-SY5Y-CRE cell line as a valid system for functionally testing fluorescent rAAV vectors, we transduced the cells with rAAV containing neuronal-specific and ubiquitous promoters, as well as, across several common serotypes (Fig. 2). We used rAAV vectors with eGFP driven by either the neuronal-specific promoters CAMKIIα and human Synapsin (hSyn) [[6], [7], [8]], or the ubiquitously expressed promoters CAG and EF1αc (core) [[9], [10], [11]] (Fig. 2A). In all experiments, the SH-SY5Y-CRE cells were transduced as follows: SH-SY5Y-CRE cells were seeded on 24 well plates at ∼1 × 104 cells/well one day prior to rAAV transduction. The next day, 1 × 109 viral genomes (vg) of AAV (MOI = 100,000) were added per well in DMEM with 5% FBS and incubated at 37 °C and 5% CO2 for 3–5 days. Media was exchanged for 1x PBS and cells were visualized on an Olympus inverted fluorescent microscope. All promoters tested showed green fluorescence in the SH-SY5Y-CRE cells similar to the parental cell line (Fig. 2A). To examine how well different rAAV serotypes can transduce the SH-SY5Y-CRE cells, we used a pAAV-EF1αc-mCherry construct (UM Vector Core) packaged in serotypes 1, 2, 5, and 8 (Fig. 2B) [12]. All serotypes tested transduced SH-SY5Y-CRE cells and showed red fluorescence. rAAV9 was also able to transduce SH-SY5Y cells (Fig. 2A).
Fig. 2.
SH-SY5Y-CRE cells can express from neuronal and ubiquitous promoters and can be transduced using different serotypes. A. SH-SY5Y cells (top panels) and SH-SY5Y-CRE cells (lower panels) infected by rAAVs express eGFP driven by different promoters. GFP driven by hSyn (4 days post infection), CAG (3 days post infection), CAMKIIa (5 days post infection) and EF1αc (4 days post infection) are shown. B. SH-SY5Y cells (top panels) and SH-SY5Y-CRE cells (lower panels) can be transduced by different rAAV serotypes. EF1αc-mCherry was packaged in rAAV1, rAAV2, rAAV5 and rAAV8 serotypes, delivered to cells, and imaged 4 days post infection. All images were taken at 20x magnification on an Olympus IX7 UV inverted microscope.
Validation of SH-SY5Y-CRE reporter cell line for Cre-dependent AAV
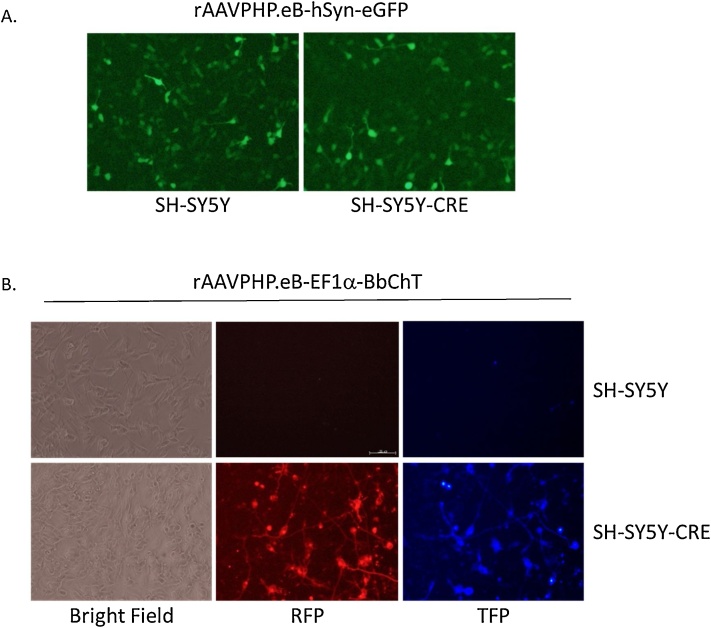
To validate SH-SY5Y-CRE cells as a rapid, cost effective functional assay for testing neuron-specific AAV constructs, we first tested the rAAV serotype PHP.eB. The rAAV-PHP.eB serotype is derived from AAV9, is able to cross the blood brain barrier to gain entry to the brain from a tail vein or retro orbital injection, and can transduce neurons in vivo [13]. Parental and SH-SY5Y-CRE cells were incubated with rAAVPHPeB-hSYN-eGFP (UM Vector Core) as described above and showed GFP expression 3 days post transduction (Fig. 3A). The rAAVPHP.eB serotype, along with rAAV-1, -2, -5, -8 and -9, is able to transduce SH-SY5Y-CRE cells.
Fig. 3.
SH-SY5Y-CRE cells are transduced by rAAV-PHP.eB and activate expression of CRE-dependent reporters. A. SH-SY5Y cells (left panel) and SH-SY5Y-CRE cells (right panel) are both transduced by rAAVPHP.eB-hSyn-eGFP. Images were taken 3 days post transduction. B. SH-SY5Y cells (top panels) and SH-SY5Y-CRE (lower panels) were transduced with the Brainbow construct rAAVPHP.eB-EF1α-BbChT. Only the SH-SY5Y-CRE cells show expression of mCherry and TFP which are CRE-dependent. Images were taken 7 days post transduction. All images were taken at 20x magnification on an Olympus IX7 UV inverted microscope.
To demonstrate that SH-SY5Y-CRE cells are an effective functional tool for testing neuron-specific floxed AAV constructs, we used the Brainbow construct pAAV-EF1a-BbChT (Addgene #45186). Brainbow vectors contain multiple floxed fluorescent proteins when recombined by Cre can give a multitude of color combinations [14]. These constructs can help identify neurons, cell lineages and cell networks. The Brainbow construct pAAV-EF1a-BbChT contains the mCherry and TFP fluorescent proteins in the reverse orientation each flanked by loxp sites where it cannot produce a functional protein [15]. In the presence of Cre, the fluorescent genes are flipped to the correct translational orientation where it is now able to be expressed and visualized. We transduced SH-SY5Y parental and Cre cells with rAAVPHPeB-EF1a-BbChT. TFP and mCherry were detected in SH-SY5Y-CRE cells but not the parental SH-SY5Y control cells 5–7 days post transduction (Fig. 3B). In this study, the SH-SY5Y-CRE cells are an effective tool as a reporter cell line for validating rAAVs with complex, Cre-dependent transgene cassettes.
Conclusion
We developed a Cre-expressing reporter cell line to assess the functional quality of Cre-dependent rAAV. We show that this SH-SY5Y-CRE neuroblastoma cell line expresses a functional Cre recombinase that can act on floxed constructs delivered by rAAV. Additionally, we show that this reporter cell line is transduced by multiple AAV serotypes, as well as, with a variety of commonly used neuronal-specific and ubiquitous promoters. These characteristics demonstrate that the SH-SY5Y-CRE cells can be used as a standard reporter cell line for a wide range of rAAV constructs.
Acknowledgements
This work was supported by the Michigan Diabetes Research Center (NIH P30 DK020572). Special thanks to the laboratories of Dr. Martin Myers and Dr. Dawen Cai for in vivo functional validation of rAAV vectors in mouse models.
Acknowledgments
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Ellis B.L., Hirsch M.L., Barker J.C., Connelly J.P., Steininger R.J., Porteus M.H. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virol. J. 2013;10:74. doi: 10.1186/1743-422X-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ross R.A., Spengler B.A., Biedler J.L. Coordinate morphological and biochemical interconversion of human neuroblastoma cells. J. Natl. Cancer Inst. 1983;71:741–747. [PubMed] [Google Scholar]
- 3.Biedler J.L., Roffler-Tarlov S., Schachner M., Freedman L.S. Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Cancer Res. 1978;38:3751–3757. [PubMed] [Google Scholar]
- 4.Teschendorf C., Warrington K.H., Jr., Siemann D.W., Muzyczka N. Comparison of the EF-1 alpha and the CMV promoter for engineering stable tumor cell lines using recombinant adeno-associated virus. Anticancer Res. 2002;22:3325–3330. [PubMed] [Google Scholar]
- 5.Qin J.Y., Zhang L., Clift K.L., Hulur I., Xiang A.P., Ren B.Z., Lahn B.T. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One. 2010;5:e10611. doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nathanson J.L., Yanagawa Y., Obata K., Callaway E.M. Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neuroscience. 2009;161:441–450. doi: 10.1016/j.neuroscience.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dittgen T., Nimmerjahn A., Komai S., Licznerski P., Waters J., Margrie T.W., Helmchen F., Denk W., Brecht M., Osten P. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc. Natl. Acad. Sci. U. S. A. 2004;101:18206–18211. doi: 10.1073/pnas.0407976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yaguchi M., Ohashi Y., Tsubota T., Sato A., Koyano K.W., Wang N., Miyashita Y. Characterization of the properties of seven promoters in the motor cortex of rats and monkeys after lentiviral vector-mediated gene transfer. Hum. Gene Ther. Methods. 2013;24:333–344. doi: 10.1089/hgtb.2012.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 10.Gray S.J., Foti S.B., Schwartz J.W., Bachaboina L., Taylor-Blake B., Coleman J., Ehlers M.D., Zylka M.J., McCown T.J., Samulski R.J. Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Hum. Gene Ther. 2011;22:1143–1153. doi: 10.1089/hum.2010.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Xu Z., Tian Z., Zhang X., Xu D., Li Q., Zhang J., Wang T. The EF-1α promoter maintains high-level transgene expression from episomal vectors in transfected CHO-K1 cells. J. Cell. Mol. Med. 2017;21:3044–3054. doi: 10.1111/jcmm.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Z., Asokan A., Samulski R.J. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol. Ther. 2006;14:316–327. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Chan K.Y., Jang M.J., Yoo B.B., Greenbaum A., Ravi N., Wu W.L., Sánchez-Guardado L., Lois C., Mazmanian S.K., Deverman B.E., Gradinaru V. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017;20:1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livet J., Weissman T.A., Kang H., Draft R.W., Lu J., Bennis R.A., Sanes J.R., Lichtman J.W. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 15.Cai D., Cohen K.B., Luo T., Lichtman J.W., Sanes J.R. Improved tools for the Brainbow toolbox. Nat. Methods. 2013;10:540–547. doi: 10.1038/nmeth.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]