Abstract
Background
Autophagy is a self-degrading process. Previously, we showed that insulin-like growth factor binding protein-related protein 1 (IGFBPrP1) is a novel transforming growth factor β1 (TGFβ1)-interacting factor in liver fibrosis; the role of TGFβ1-mediated autophagy in hepatic stellate cells (HSCs) activation has been investigated. However, whether autophagy is regulated by IGFBPrP1 remains unknown.
Aims
We investigated the interactions among IGFBPrP1, autophagy, and activation of primary rat HSCs.
Methods
Primary HSCs were separated from Sprague Dawley rats by two-step enzymatic digestion, and then, we overexpressed or inhibited IGFBPrP1 expression in HSCs under serum-starved condition. Autophagy inducer rapamycin or inhibitor 3-methyladenine (3MA) was used to assess the relationship between autophagy and HSCs activation.
Results
We observed the expression of activation marker α-SMA and autophagy markers such as LC3B and Beclin1, which were significantly increased in HSCs treated with adenovirus vector harboring the IGFBPrP1 gene (AdIGFBPrP1) compared to cells cultured under serum-starved. In comparison, HSCs treated with shIGFBPrP1 showed opposite results. Furthermore, HSCs activation and autophagy increased when cells were treated with rapamycin, whereas opposite results were obtained when cells were treated with 3MA. AdIGFBPrP1 treatment downregulated the phosphorylation of Akt and mTOR.
Conclusion
Autophagy was induced in IGFBPrP1-treated primary HSCs, and IGFBPrP1-induced autophagy promoted the activation of HSCs and extracellular matrix expression, the underlying mechanism of which may involve the phosphatidylinositide 3-kinase/Akt/mTOR signaling pathway.
Keywords: IGFBPrP1, Autophagy, Liver fibrosis, PI3K/Akt/mTOR signaling pathway, Hepatic stellate cells
Introduction
Liver fibrosis is a progressive pathological process, which is the result of increased expression of extracellular matrix (ECM) and reduced degradation of collagen fibers [1]. Liver fibrosis and cirrhosis are challenging medical problems, and therefore, studies on the development of new therapeutic strategies or targets are of considerable value. Autophagy is an endocellular catabolic mechanism via which cytoplasmic proteins and organelles are degraded by lysosomes for maintaining cellular homeostasis [2]. Autophagy is associated with several diseases [3–6], including liver disease. He et al. [7] observed that LC3 expression increased, whereas that of SQSTM1/p62 decreased following activation of hepatic stellate cells (HSCs) isolated from rats with liver fibrosis. One study [8] showed that autophagy releases lipids which could promote fibrogenesis by activated HSCs in mice and human tissues. Another study demonstrated that inhibition of autophagy could reverse alcohol-induced HSCs activation [9]. While evidences support the theory that autophagy is associated with liver fibrosis and HSCs activation, the underlying molecular mechanisms are complex and elusive.
Owing to hepatocyte injury, necrosis, and immune response activation, factors such as sinusoidal endothelial cells, hepatocytes, Kupffer cells, and HSCs are involved in the development of hepatic fibrosis [10, 11]. HSCs activation is the key for promoting liver fibrosis, and various cytokines participate in this process [12]. Transforming growth factor β1 (TGFβ1) is an important profibrotic cytokine that promotes fibroblast recruitment, proliferation, differentiation into myofibroblasts, and ECM production [13]. The insulin-like growth factor binding protein-related protein 1 (IGFBPrP1), also known as the insulin-like growth factor binding protein 7 (IGFBP7), is a new TGFβ1-interacting profibrotic cytokine. We previously showed that IGFBPrP1 overexpression promoted the expression of TGFβ1 and ECM in vitro and vivo [14, 15]. Moreover, we observed that overexpression of TGFβ1 increased IGFBPrP1 levels with HSCs activation. Similarly, overexpression of IGFBPrP1 activated HSCs and upregulated TGFβ1 [16].
Whether TGFβ1 regulates autophagy during HSCs activation has been investigated. TGFβ1 induced autophagy flux in primary rat HSCs [17], protected HSC-T6 from serum deprivation, and reduced apoptosis via autophagy activation [18]. However, whether IGFBPrP1 regulates autophagy is not yet clear. Autophagy is regulated by multiple signaling pathways; PI3K/Akt/mTOR signaling pathway is particularly critical [19]. IGFBPrP1 has been shown to inhibit insulin signaling in vitro [20]. One study found that pretreatment of normal and breast cancer cells with IGFBPrP1 induced the accumulation of inactive IGF1R on the cell surface and blockade of downstream PI3K/Akt signaling [21]. Another study found that ConA-induced liver fibrosis and autophagy are mediated by the PI3K/Akt signaling pathway; the protein levels of PI3K and phosphorylated Akt were downregulated [22]. Thus, we hypothesized that IGFBPrP1 may modulate autophagy through PI3K/Akt/mTOR signal pathway during HSCs activation.
In the present study, primary rats HSCs were used as their biological characters were not significantly altered and they closely mimicked the in vivo cellular state compared to HSCs line. We detected autophagy markers such as Beclin1 in the initial stage, LC3B in the formation stage, and the autophagic degradation substrate SQSTM1/p62 during the multi-step process of autophagy. Thus, the aim of this study was to investigate the effect of IGFBPrP1 stimulation on autophagy and primary HSCs activation, and the relationship between them.
Methods
Primary Cell Isolation, Culture, and Identification
Animals were obtained from Shanxi Medical University Laboratory Animal Center (Taiyuan, China). Healthy male Sprague Dawley rats were anesthetized by intraperitoneal injection of 10% chloral hydrate, their livers were perfused and digested with type IV collagenase via the portal vein, and primary HSCs were separated and purified using Nycodenz. Cell viability was determined by trypan blue staining. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Biological Industries, USA) supplemented with 10% fetal bovine serum (FBS; Biological Industries) and 100 U/ml penicillin/streptomycin. The cells were incubated at 37 °C with 5% CO2 in a humidified incubator, the medium was replaced after every two days, and 3–4 generations of cells were used as work cells. Lipid droplets’ presence was visualized by microscopy during the established time of culture. Antibodies against desmin (TransGen Biotech, Beijing, China) and α-smooth muscle actin (α-SMA) (Abcam, Cambridge, UK) were used to identify HSCs.
Cell Transfection and Treatment
Primary HSCs were transfected with adenovirus vector harboring the IGFBPrP1 gene (AdIGFBPrP1) (Gene Pharma Company, Shanghai, China), at a multiplicity of infection (MOI) of 10, 20, 40, or 80 (number of viruses/number of cells). The transfection efficacy was evaluated by detecting the number of EGFP-positive cells. The optimized MOI80 was used in subsequent experiments. Three short hairpin RNAs (shRNAs) targeting the rat IGFBPrP1 mRNA were designed and synthesized (Gene Pharma Company, Shanghai, China). The most effective shIGFBPrP1 was used in subsequent experiments. The primary cells were treated with a gradient dose and time course of autophagy inducer rapamycin (Solarbio Company, Beijing, China) or inhibitor 3MA (Solarbio Company, Beijing, China) under serum starvation condition to determine the proper time and dosage required for the subsequent experiments. The primary cells were treated with chloroquine (30 μM) for 24 h.
Transmission Electron Microscopy (TEM)
The treated cells were collected by centrifugation, followed by fixing first with 2.5% glutaraldehyde for 2 h at 4 °C and then with 1% osmium tetroxide for 1 h at 4 °C. The samples were dehydrated in a graded a series of ethanol baths, infiltrated, and embedded in EPON resin. Finally, the samples were cut into ultrathin sections of 50 nM thickness, double stained with uranyl acetate and lead citrate, and observed under a transmission electron microscope (Hitachi, Tokyo, Japan).
Autophagy Flux Detection
Primary HSCs were cultured in 24-well plates (3 × 105 cells/well) and treated with Ad-mRFP-GFP-LC3 (HanBio, Wuhan, China) at 40 MOI. The cells were washed with PBS, fixed with 4% paraformaldehyde, and observed under a laser confocal microscopy (Nikon, Tokyo, Japan). The number of GFP and mRFP dots was determined by counting of fluorescence puncta from 30 different cells. The number of dots per cell was calculated by dividing the total number of dots by the number of cells in each microscopic field.
MDC Staining
Primary HSCs were cultured in 24-well plates (3 × 105 cells/well). Cells were incubated with MDC (50 μM) (Solarbio Life Science, Beijing, China) at 37 °C for 1 h in the dark. After incubation, the cells were washed twice with phosphate-buffered saline (PBS), and fluorescent micrographs were observed under a fluorescence microscope.
Reverse Transcription Quantitative Polymerase Chain Reaction (qPCR)
Total RNA was extracted from cells using an Eastep™ Super Total RNA Extraction Kit (Promega, Madison, USA) following the manufacturer’s instructions. RNA was reverse-transcribed using the GoScript™ Reverse Transcription Mix (Promega, Madison, USA). GoTaq® qPCR Master Mix (Promega, Madison, USA), cDNA template, and primers were mixed in a volume of 20 μL using the Step One Real-Time PCR System (Applied Biosystems, Foster City, USA). The primer sequences were as follows:
| IGFBPrP1 | Forward | 5′-GAAGTAACTGGCTGGGTGCT-3′ |
| Reverse | 5′-AATTTTGGCCGACGCTGAAG-3′ | |
| TGFβ1 | Forward | 5′-CACTCCCGTGGCTTCTAGTG-3′ |
| Reverse | 5′-CTGGCGAGCCTTAGTTTGGA-3′ | |
| LC3 | Forward | 5′-CAGGACAAGCAGGCAGATGA-3′ |
| Reverse | 5′-GGCTTTCGTCTCTTCCACCA-3′ | |
| α-SMA | Forward | 5′-GGCTCTGGGCTCTGTAAGG-3′ |
| Reverse | 5′-CTCTTGCTCTGGGCTTCATC-3′ | |
| GAPDH | Forward | 5′-GCGAGATCCCGCTAACATCA-3′ |
| Reverse | 5′-CTCGTGGTTCACACCCATCA-3′ |
The data were analyzed using the ΔΔ threshold (Ct) method.
Western Blotting
Total protein was obtained from cells using a Total Protein Extraction Kit (KeyGEN BioTECH, Jiangsu, China) following the manufacturer’s protocol. Equal amounts of samples were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA). The primary antibodies used were as follows: IGFBPrP1, TGFβ1, α-SMA, collagen I, LC3B, Beclin1, SQSTM1/p62, mTOR, p-mTOR, Akt, p-Akt (Abcam, Cambridge, UK), and β-actin (TransGen Biotech, Beijing, China). Horseradish peroxidase (HRP)-conjugated IgG was used as the secondary antibody. Protein bands were detected using a super-enhanced chemiluminescence (ECL) detection kit (Amersham Pharmacia Biotech, NJ, USA).
Statistical Analysis
Statistical analysis was performed using the SPSS software, version 19.0. Results were presented as mean ± standard deviation (SD). Analysis of variance (ANOVA) and Mauchly or Greenhouse–Geisser tests were used to compare repeated measured data, and P ≤ 0.05 was considered statistically significant.
Results
Effect of AdIGFBPrP1 on α-SMA, Collagen I, IGFBPrP1, and TGFβ1 Expression Levels in Primary HSCs
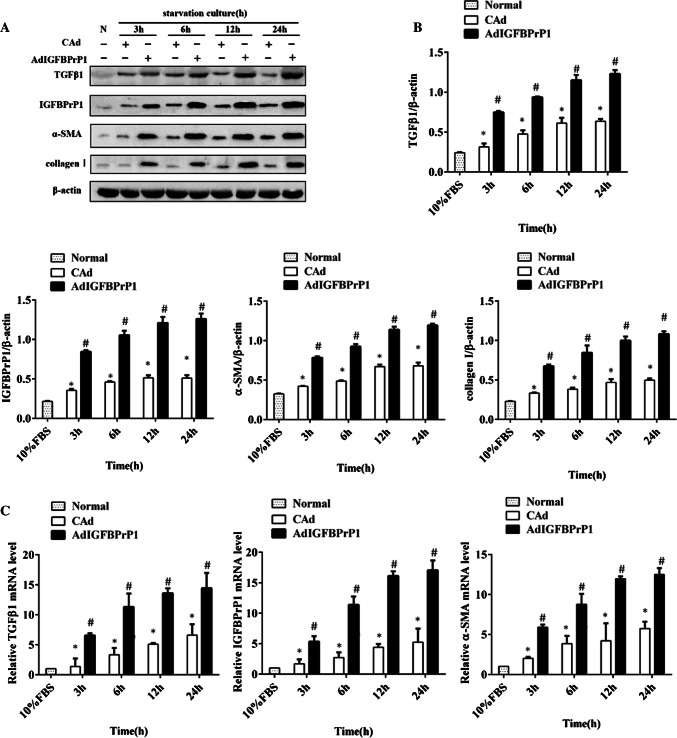
To determine changes in the levels of activation and ECM expression in the AdIGFBPrP1-transfected HSCs with time, we assessed the protein and mRNA levels of α-SMA, collagen I, IGFBPrP1, and TGFβ1 by Western blotting and qPCR. Following treatment with AdIGFBPrP1, the cells showed an increase in IGFBPrP1 levels, which peaked at 24 h, indicating the efficiency of AdIGFBPrP1 transfection (Fig. 1a–c). The TGFβ1 protein (Fig. 1a, b) and mRNA levels (Fig. 1c) gradually increased from 3 to 24 h. In addition, both α-SMA and collagen I expressions increased at 3 h, followed by a gradual increase (Fig. 1a–c).
Fig. 1.
Overexpression of IGFBPrP1-induced activation and ECM expression in primary HSCs. a Cells were cultured in 10% FBS or serum starvation medium with AdIGFBPrP1 or adenoviral vectors carrying no cDNA (CAd) for various time periods, and protein levels of α-SMA, collagen I, IGFBPrP1, and TGFβ1 were analyzed by Western blotting. b Band intensities of α-SMA, collagen I, IGFBPrP1, and TGFβ1 relative to the control cells were determined after normalization to β-actin expression. c mRNA levels of α-SMA, IGFBPrP1, and TGFβ1 were determined using qPCR. *P< 0.05 compared to the normal control (10% FBS). #P< 0.05 compared to the CAd control (2% FBS). Data were presented as mean ± SD for three replicate experiments (n = 3 per group)
Effect of AdIGFBPrP1 on Beclin1, LC3B, and SQSTM1/p62 Expression Levels in Primary HSCs
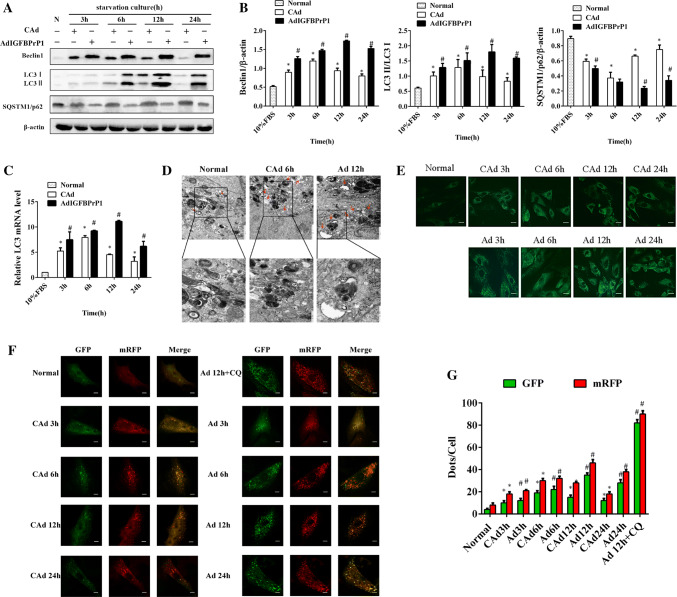
To determine the levels of autophagy in the AdIGFBPrP1-transfected HSCs, we assessed the protein levels of Beclin1, SQSTM1/p62, and LC3B by Western blot analysis (Fig. 2a, b). When HSCs were treated with AdIGFBPrP1 at different time points, the protein levels of Beclin1 and the LC3 II/LC3 I ratio increased in 3 h, peaked at 12 h, and then decreased gradually with time. AdIGFBPrP1 treatment increased Beclin1 and LC3B protein levels to a larger extent than serum starvation. In contrast, the protein levels of SQSTM1/p62 showed an opposite trend. As shown in Fig. 2c, the LC3B mRNA levels of the AdIGFBPrP1 groups increased at 3, 6, and 12 h, which peaked at 12 h.
Fig. 2.
Overexpression of IGFBPrP1 induced autophagy in primary HSCs. a Cells were cultured in 10% FBS or serum starvation medium with AdIGFBPrP1 or CAd for various time periods, and the protein levels of Beclin1, LC3B, and SQSTM1/p62 were analyzed by Western blotting. b Band intensities of Beclin1, LC3B, and SQSTM1/p62 relative to the control cells were determined after normalizing to β-actin expression. c mRNA levels of LC3B were measured by qPCR. d Cells were cultured in serum starvation medium with CAd for 6 h or AdIGFBPrP1 for 12 h. Images were acquired by TEM. Scale bar, 1 μm and 500 nm. e AVO detection after MDC staining by fluorescence microscopy. Scale bar, 20 μm. f Cells were transfected with mRFP-GFP-LC3 adenovirus and assessed using confocal microscopy. Scale bar, 50 μm. g Mean numbers of GFP and mRFP dots per cell. *P< 0.05 compared to the normal control (10% FBS). #P< 0.05 compared to the CAd control (2% FBS). Data were presented as mean ± SD for three replicate experiments (n = 3 per group)
As TEM is the gold standard for identifying autophagy, ultrastructural changes in treated cells were examined using TEM. TEM images of control cells showed normal cytoplasm and were characterized by mitochondria, endoplasmic reticulum, free ribosomes, lysosomes, and few autophagosomes (Fig. 2d). In contrast, the AdIGFBPrP1-treated cells exhibited numerous autophagosomes and autolysosomes in the cytoplasm. The serum-starved cells with CAd exhibited less autophagosomes and autolysosomes than the AdIGFBPrP1-treated cells. MDC, a fluorescent substance, is a specific marker for cytoplasmic acidic vascular organelles (AVOs), including autolysosomes and autophagosomes, and was used to detect autophagy [23]. We observed that HSCs overexpressing IGFBPrP1 showed greater fluorescence intensity and more cytoplasmic AVO formation than the serum-starved groups with CAd (Fig. 2e), indicating that overexpression of IGFBPrP1 increased MDC recruitment to autolysosomes. We further detected autophagy flux by using adenovirus harboring tandem fluorescent mRFP-GFP-LC3. Next, the number of autophagosomes can be evaluated by determining the number of red dots that overlay green dots and appear yellow in merged images. The red dots that do not overlay green dots and appear red in merged images indicate autolysosome formation. The numbers of green and red dots per cell were both significantly increased in the IGFBPrP1-treated groups compared to the serum-starved groups with CAd (Fig. 2f, g), which was similar to the results obtained in Western blot analysis.
Effect of Rapamycin on α-SMA, Collagen I, IGFBPrP1, and TGFβ1 Levels in AdIGFBPrP1-Treated Primary HSCs
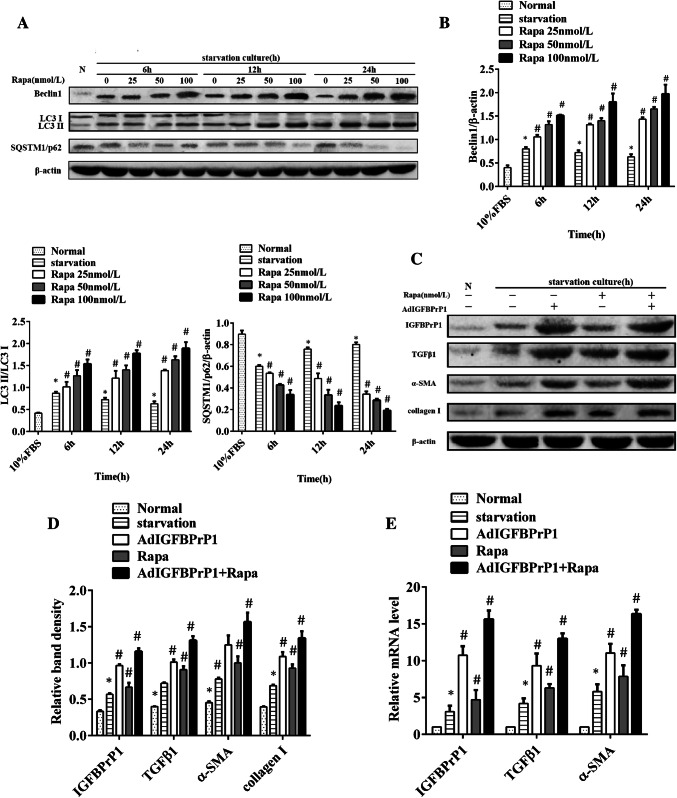
To determine the proper time and dosage required for observing the effect of rapamycin, we treated the HSCs with a gradient dose course of rapamycin ranging from 25 to 100 nmol/L and a gradient time course ranging from 6 to 24 h. Western blotting was used to determine autophagy levels (Fig. 3a, b). Consistent with the fact that increase in Beclin1 level and LC3II/LC3I ratio and decrease in SQSTM1/p62 levels are credible markers of autophagy enhancement, our results showed that rapamycin promoted Beclin1 and LC3B expression and inhibited SQSTM1/p62 expression in a time- and dose-dependent manner.
Fig. 3.
Rapamycin increases IGFBPrP1-induced activation and ECM expression in primary HSCs. a HSCs were treated with rapamycin in a gradient dose and time course. Western blotting was used to analyze the expression levels of Beclin1, SQSTM1/p62, and LC3B. b Band intensities of Beclin1, SQSTM1/p62, and LC3B relative to the control cells were determined after normalization to β-actin expression. c HSCs were cultured in 10% FBS or 2% FBS with or without AdIGFBPrP1 and rapamycin (100 nmol/L) for 24 h. The protein levels of α-SMA, collagen I, IGFBPrP1, and TGFβ1 were analyzed by Western blotting. d Band intensities of α-SMA, collagen I, IGFBPrP1, and TGFβ1 relative to the control cells were determined after normalization to β-actin expression. e The mRNA levels of α-SMA, IGFBPrP1, and TGFβ1 were measured by qPCR. *P< 0.05 compared to the normal control (10% FBS). #P< 0.05 compared to the serum starvation (2% FBS). Data were presented as mean ± SD for three replicate experiments (n = 3 per group)
The effect of rapamycin on HSCs activation and ECM expression induced by AdIGFBPrP1 is shown in Fig. 3c–e. Cells which were cultured in serum starvation medium with both AdIGFBPrP1 and rapamycin showed higher protein and mRNA levels of IGFBPrP1 and TGFβ1 than other groups where HSCs were cultured with either AdIGFBPrP1 or rapamycin. In addition, the expression of α-SMA and collagen I was also increased. These results indicated that rapamycin promoted IGFBPrP1 and TGFβ1 expression and induced HSCs activation and ECM expression via AdIGFBPrP1.
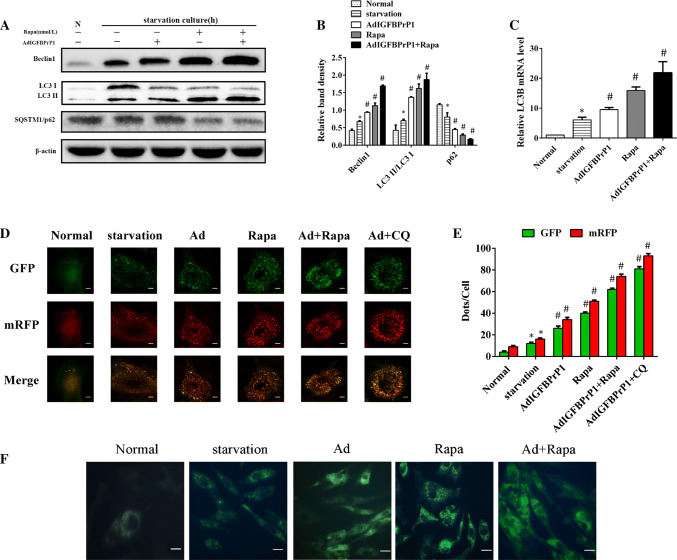
Effect of Rapamycin on Beclin1, LC3B, and SQSTM1/p62 Expression in AdIGFBPrP1-Treated Primary HSCs
To investigate the effect of rapamycin on autophagy of IGFBPrP1-treated HSCs, cells were cultured in serum-starved medium with or without IGFBPrP1 or rapamycin for 24 h. The ratio of LC3I to LC3II conversion, Beclin1 upregulation, and SQSTM1/P62 degradation indicated that rapamycin significantly promoted IGFBPrP1-induced autophagy in HSCs (Fig. 4a, b). Similarly, the mRNA levels (Fig. 4c) of LC3B were also upregulated after rapamycin and IGFBPrP1 treatment. We used mRFP-GFP-LC3 to further verify this phenomenon (Fig. 4d, e) and observed that the numbers of green and red dots per cell which were both significantly increased in rapamycin and IGFBPrP1-treated groups were higher than those of other groups. In addition, the same phenomenon was observed in MDC staining (Fig. 4f).
Fig. 4.
Rapamycin increases IGFBPrP1-induced autophagy in primary HSCs. a HSCs were cultured in 10% FBS or 2% FBS with or without AdIGFBPrP1 and rapamycin (100 nmol/L) for 24 h. The protein levels of Beclin1, SQSTM1/p62, and LC3B were analyzed by Western blotting. b Band intensities of Beclin1, SQSTM1/p62, LC3B relative to the untreated control cells were determined after normalization to β-actin expression. c mRNA levels of LC3B were measured by qPCR. d Cells were transfected with mRFP-GFP-LC3 adenovirus and assessed using confocal microscopy. Scale bar, 50 μm. e Mean numbers of GFP and mRFP dots per cell. f Autophagosome formation was observed by MDC staining. Scale bar, 20 μm. *P< 0.05 compared to the normal control (10% FBS). #P< 0.05 compared to the serum starvation (2% FBS). Data were presented as mean ± SD for three replicate experiments (n = 3 per group)
Effect of 3MA on α-SMA, Collagen I, IGFBPrP1, and TGFβ1 Expression in AdIGFBPrP1-Treated Primary HSCs
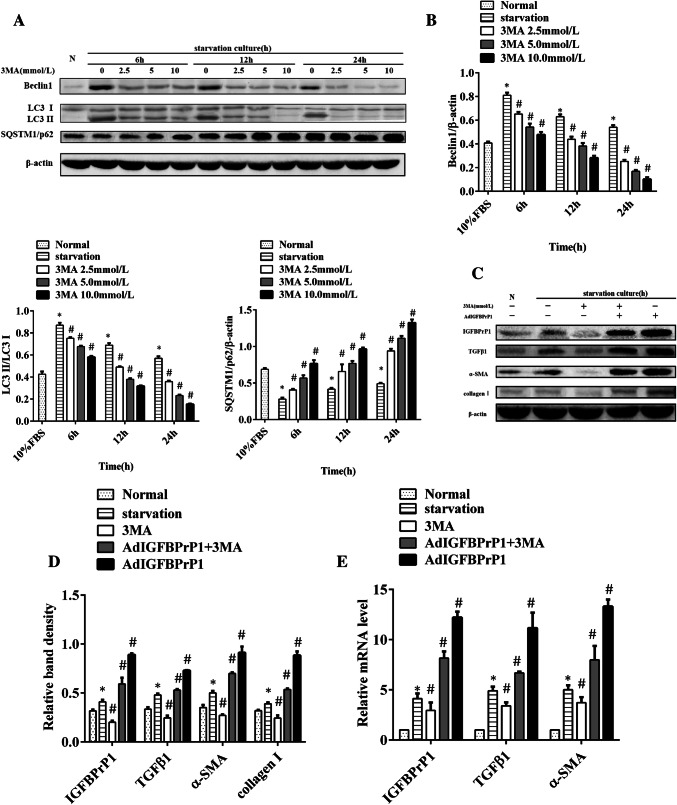
We treated HSCs with the autophagy inhibitor 3MA and set gradient dose and time courses ranging from 2.5 to 10.0 mmol/L and 6 to 24 h, respectively. Western blotting was used to examine autophagy levels to select the proper time and dosage required for observing the effect of 3MA (Fig. 5a, b). The decrease in Beclin1 level and LC3II/LC3I ratio and increase in SQSTM1/p62 level showed that 3MA reduced Beclin1 and LC3B expression and promoted SQSTM1/p62 expression in a time- and dose-dependent manner.
Fig. 5.
3MA decreases IGFBPrP1-induced activation and ECM expression in primary HSCs. a HSCs were treated with 3MA in a gradient dose and time course. Western blotting was used to analyze the expression levels of Beclin1, SQSTM1/p62, and LC3B. b Band intensities of Beclin1, SQSTM1/p62, and LC3B relative to the untreated control cells were determined after normalization to β-actin expression. c HSCs were cultured in 10% FBS or 2% FBS with or without AdIGFBPrP1 and 3MA (10.0 mmol/L) for 24 h. α-SMA, collagen I, IGFBPrP1, and TGFβ1 levels were analyzed by Western blotting. d Band intensities of α-SMA, collagen I, IGFBPrP1, and TGFβ1 relative to the untreated control cells were determined after normalization to β-actin expression. e mRNA levels of α-SMA, IGFBPrP1, and TGFβ1 were determined by qPCR. *P< 0.05 compared to the normal control (10% FBS). #P< 0.05 compared to the serum starvation (2% FBS). Data were presented as mean ± SD for three replicate experiments (n = 3 per group)
The effect of 3MA on IGFBPrP1-induced HSCs activation and ECM expression is shown in Fig. 5c–e. HSCs cultured in serum starvation medium with both IGFBPrP1 and 3MA showed lower protein and mRNA levels of IGFBPrP1 and TGFβ1 than those of cells cultured only with IGFBPrP1. In addition, α-SMA and collagen I expression was also reduced. These results indicated that 3MA inhibited the expression of IGFBPrP1 and TGFβ1 and further inhibited HSCs activation and ECM expression induced by IGFBPrP1.
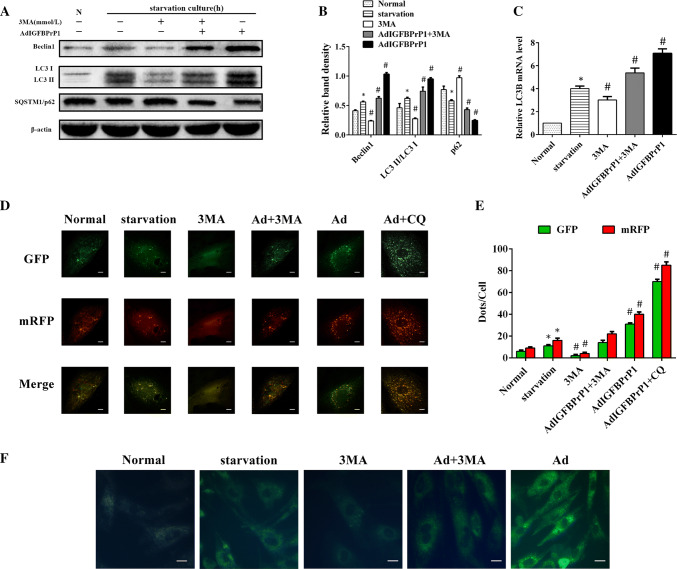
Effect of 3MA on Beclin1, LC3B, and SQSTM1/p62 Expression in AdIGFBPrP1-Treated Primary HSCs
HSCs were cultured in serum starvation medium with or without IGFBPrP1 or 3MA for 24 h. The ratio of LC3I to LC3II conversion and Beclin1 level decreased, whereas SQSTM1/p62 level increased, which indicated that 3MA inhibited IGFBPrP1-induced autophagy in HSCs (Fig. 6a, b). 3MA treatment also reduced LC3B mRNA levels (Fig. 6c). In addition, the numbers of green and red dots per cell of 3MA and IGFBPrP1-treated groups were lesser than those of the IGFBPrP1-treated groups (Fig. 6d, e). Besides, the least fluorescence intensities of 3MA and IGFBPrP1-treated groups were observed in MDC staining (Fig. 6f). Together, these results indicated that 3MA inhibited IGFBPrP1-induced autophagy in primary HSCs.
Fig. 6.
3MA decreases IGFBPrP1-induced autophagy in primary HSCs. a HSCs were cultured in 10% FBS or 2% FBS with or without AdIGFBPrP1 and 3MA (10.0 mmol/L) for 24 h. Beclin1, SQSTM1/p62, and LC3B levels were analyzed by Western blotting. b Band intensities of Beclin1, SQSTM1/p62, and LC3B relative to the untreated control cells were determined after normalization to β-actin expression. c mRNA levels of LC3B were measured by qPCR. d Cells were transfected with mRFP-GFP-LC3 adenovirus and assessed using confocal microscopy. Scale bar, 50 μm. e Mean numbers of GFP and mRFP dots per cell. f Autophagosome formation was observed by MDC staining. Scale bar, 20 μm. *P< 0.05 compared to the normal control (10% FBS). #P< 0.05 compared to the serum starvation (2% FBS). Data were presented as mean ± SD for three replicate experiments (n = 3 per group)
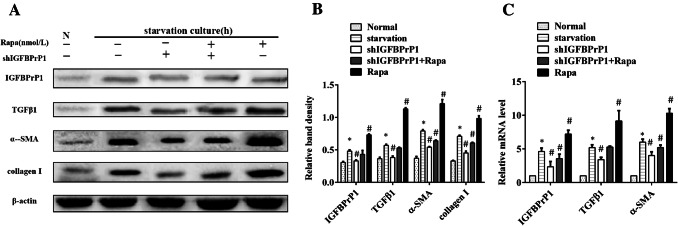
Effect of shIGFBPrP1 on α-SMA, Collagen I, IGFBPrP1, and TGFβ1 Expression Levels in Primary HSCs
To investigate the effect of shIGFBPrP1 on activation and ECM expression of serum starvation and rapamycin-treated HSCs, cells were cultured in serum-starved medium with or without shIGFBPrP1 or rapamycin for 24 h. As shown in Fig. 7a–c, cells which were cultured in serum starvation medium with shIGFBPrP1 showed lower protein and mRNA levels of IGFBPrP1 and TGFβ1 than other groups where HSCs were cultured under serum starvation medium. Besides, the expression of α-SMA and collagen I was also decreased. HSCs cultured in serum starvation medium with both shIGFBPrP1 and rapamycin showed same results than those of cells cultured only with rapamycin. These results indicated that shIGFBPrP1 inhibited serum starvation or rapamycin induced HSCs activation and ECM expression.
Fig. 7.
ShIGFBPrP1 decreases serum starvation, and rapamycin induced activation and ECM expression in primary HSCs. a HSCs were cultured in 10% FBS or serum starvation medium with or without shIGFBPrP1 and rapamycin (100 nmol/L) for 24 h. α-SMA, collagen I, IGFBPrP1, and TGFβ1 levels were analyzed by Western blotting. b Band intensities of α-SMA, collagen I, IGFBPrP1, and TGFβ1 relative to the untreated control cells were determined after normalization to β-actin expression. c mRNA levels of α-SMA, IGFBPrP1, and TGFβ1 were determined by qPCR. *P< 0.05 compared to the normal control (10% FBS). #P< 0.05 compared to the serum starvation (2% FBS). Data were presented as mean ± SD for three replicate experiments (n = 3 per group)
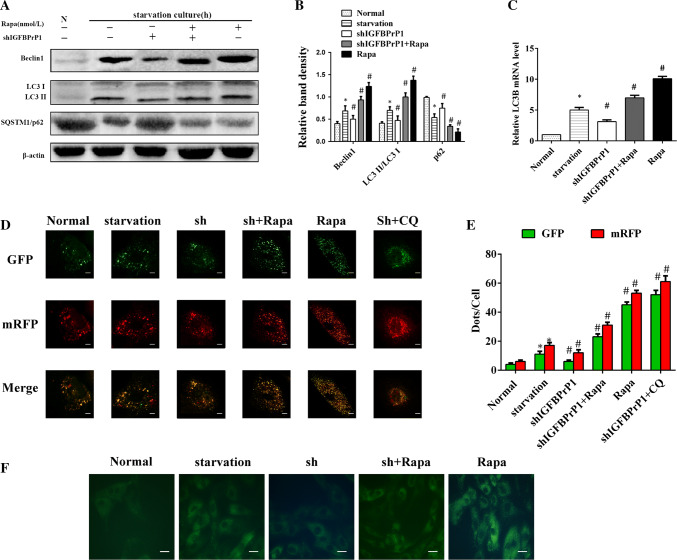
Effect of shIGFBPrP1 on Beclin1, LC3B, and SQSTM1/p62 Expression Levels in Primary HSCs
To determine the levels of autophagy in the shIGFBPrP1-transfected HSCs under serum starvation, we assessed the protein levels of Beclin1, SQSTM1/p62, and LC3B by Western blot analysis (Fig. 8a, b). The ratio of LC3I to LC3II conversion, Beclin1 downregulation, and SQSTM1/P62 upregulation indicated that shIGFBPrP1 significantly blocked serum starvation and rapamycin induced autophagy in HSCs. Similarly, the mRNA levels of LC3B were also downregulated after shIGFBPrP1 treatment (Fig. 8c). We also used mRFP-GFP-LC3 and MDC staining to further verify this phenomenon (Fig. 8d–f); the numbers of green and red dots per cell in shIGFBPrP1 treated groups were lower than those of other groups treated under serum starvation medium with or without rapamycin. The MDC staining showed same results.
Fig. 8.
ShIGFBPrP1 decreases serum starvation, and rapamycin induced autophagy in primary HSCs. a HSCs were cultured in 10% FBS or serum starvation medium with or without shIGFBPrP1 and rapamycin (100 nmol/L) for 24 h. Beclin1, SQSTM1/p62, and LC3B levels were analyzed by Western blotting. b Band intensities of Beclin1, SQSTM1/p62, and LC3B relative to the untreated control cells were determined after normalization to β-actin expression. c mRNA levels of LC3B were determined by qPCR. d Cells were transfected with mRFP-GFP-LC3 adenovirus and assessed using confocal microscopy. Scale bar, 50 μm. e Mean numbers of GFP and mRFP dots per cell. f Autophagosome formation was observed by MDC staining. Scale bar, 20 μm. *P< 0.05 compared to the normal control (10% FBS). #P< 0.05 compared to the serum starvation (2% FBS). Data were presented as mean ± SD for three replicate experiments (n = 3 per group)
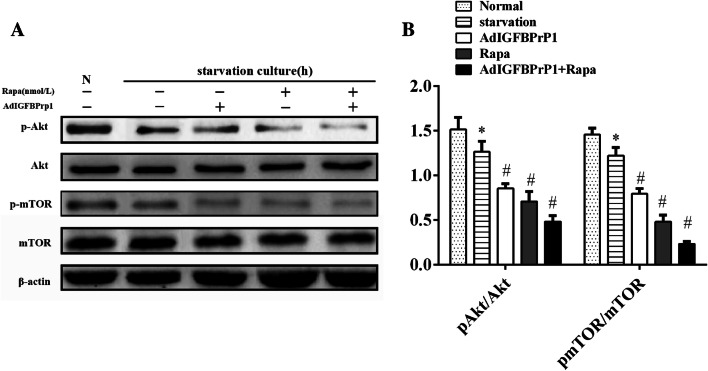
AdIGFBPrP1-Induced Autophagy Is Mediated by the PI3K/Akt/mTOR Signaling Pathway
To determine whether the PI3K/Akt/mTOR signaling pathway regulates IGFBPrP1-induced autophagy and activation, Akt, p-Akt, mTOR, and p-mTOR levels were analyzed by Western blotting (Fig. 9a, b). AdIGFBPrP1 treatment reduced p-Akt and p-mTOR levels, which indicated that IGFBPrP1 might promote HSCs activation by mediating the phosphorylation of Akt and mTOR and reducing the upstream kinase activity of mTOR to inhibit autophagy.
Fig. 9.
Expression of the PI3K/Akt/mTOR signaling pathway components in HSCs. a HSCs were cultured in 10% FBS or serum starvation medium with or without AdIGFBPrP1, rapamycin (100 nmol/L), and 3MA (10.0 mmol/L) for 24 h. Akt, p-Akt, mTOR, and p-mTOR levels were analyzed by Western blotting. b Band intensities Akt, p-Akt, mTOR, and p-mTOR relative to the untreated control cells were determined after normalization to β-actin expression. *P< 0.05 compared to the normal control (10% FBS). Data were presented as mean ± SD (n = 3 per group)
Discussion
IGFBPrP1, a profibrotic factor, plays an important role in liver fibrosis. Previously, we observed that IGFBPrP1 activated quiescent HSCs, increased α-SMA expression, and caused excessive ECM expression [24, 25]. At different time points after AdIGFBPrP1 transfection, cells showed an increase in IGFBPrP1 protein and mRNA levels, which indicated that transfection was successful. Furthermore, we showed that α-SMA expression increased gradually in a time-dependent manner in primary HSCs treated with AdIGFBPrP1. ECM components change from normal basement matrix components to a fibrotic matrix, which includes collagen I [26]. Collagen I expression was also upregulated gradually by AdIGFBPrP1. These results are consistent with earlier results, which indicated that overexpression of IGFBPrP1 induced HSCs activation and excessive ECM expression. Previously, we demonstrated a mutual regulation between IGFBPrP1 and TGFβ1, where IGFBPrP1 acts as an upstream regulatory factor of TGFβ1 in liver fibrosis [16]. In this study, we observed that the mRNA and protein levels of TGFβ1 gradually increased after AdIGFBPrP1 treatment, which corroborates our previous results.
Several studies have highlighted the importance of TGFβ1 in the regulation of autophagy. One study showed that TGFβ1 induced autophagy and enhances fibrogenesis in primary cardiac fibroblasts [27]. Another study showed that TGFβ1 protected annulus fibrosus cells under serum starvation from apoptosis by downregulating excessive autophagy [28]. As autophagy is a dynamic process, several aspects of autophagy were analyzed, such as cell morphology and protein and mRNA levels of autophagic markers. TEM revealed that large numbers of autophagosomes and autolysosomes were present in HSCs cultured in serum starvation with AdIGFBPrP1. Among the three different subtypes of LC3, namely A, B, and C, LC3B is commonly used to mark autophagosomes. During autophagy, the cytosolic form of LC3 (LC3 I) is conjugated to phosphatidylethanolamine to form LC3-phosphatidylethanolamine conjugate (LC3 II), which is recruited to autophagosomal membranes [29]. Western blot analysis of LC3 II/LC3 I showed a significant conversion of LC3-I to LC3-II after AdIGFBPrP1 treatment, and the expression peaked at 12 h. The qPCR of LC3B yielded similar results. Beclin1, a part of the class III PI3K lipid kinase complex, positively affects autophagy [30]. High Beclin1 levels in AdIGFBPrP1-treated HSCs indicated that IGFBPrP1-induced autophagy may be associated with the class III PI3K complex. SQSTM1/p62 is an autophagic substrate, the expression of which is inversely related to autophagic flow [31]. In this study, we observed that the protein levels of SQSTM1/p62 showed an opposite trend to those of LC3B and Beclin1. Based on the above results, we concluded that IGFBPrP1 promoted autophagy and activated primary rat HSCs under serum starvation conditions.
Reports show that autophagy, which is mainly a cellular protective mechanism, degrades proteins and intracellular structures in response to stress, with activation of HSCs. To determine whether IGFBPrP1-induced autophagy activated HSCs, we cultured AdIGFBPrP1-transfected HSCs in serum starvation medium with rapamycin or 3MA for 24 h. Rapamycin is a prototypical promoter of autophagy, which is the inhibitor of mechanistic target of rapamycin (mTOR) complex 1 (mTORC1). mTORC1 suppresses autophagy by phosphorylating ULK1 [32]. Xie et al. [9] found that alcohol treatment increased autophagy presenting decreased p62 level and increased LC3-II/LC3-I; meantime, the expressions of α-SMA and collagen I were obviously promoted, while the above effects were strongly increased with rapamycin processing. Chen et al. [33] found that AICAR, which is a kind of AMPK activator, inhibited the expression of LC3-II and a-SMA, concomitant with significantly increased the expression of p62 in human hepatic stellate cell line LX-2, which effect could be partly reversed by treatment with the autophagy inducer rapamycin. Wu et al. [34] showed that rapamycin upregulated connective tissue growth factor expression at the transcriptional level in hepatic progenitor cells, which is a matricellular protein strongly upregulated in fibrotic liver tissue. We observed that rapamycin promoted autophagy in a time- and dose-dependent manner. These results suggested that rapamycin has potential fibrotic effect in liver. We investigated the effect of rapamycin on primary IGFBPrP1-treated HSCs. The fluorescence intensity of LC3B and AVOs was enhanced after rapamycin treatment. Furthermore, Western blotting indicated that the expression of autophagy-related proteins and mRNAs was upregulated in HSCs treated with AdIGFBPrP1 and rapamycin for 24 h compared to those in HSCs treated with AdIGFBPrP1 or rapamycin alone. Levels of collagen I and α-SMA, markers of collagen expression and HSCs activation, were increased as mentioned above. In addition, rapamycin also promoted the expression of IGFBPrP1 and TGFβ1, which suggested that rapamycin promoted the effect of IGFBPrP1 on HSCs activation by upregulating autophagy.
3MA is a pharmacological autophagy inhibitor that inhibits both class III PI3K and class I PI3K. Interestingly, the inhibitory effect of 3MA on these two autophagic targets follows opposite pattern. 3MA temporally inhibits class III PI3K, followed by inhibition of autophagy, whereas its suppressive effect on class I PI3K is persistent and promotes autophagy. Wu et al. [35] observed that cells treated with 3MA in full serum medium for long durations (up to 9 h) showed increased expression of autophagic markers. Zhao et al. [36] also observed that prolonged (24, 48, or 96 h) treatment with 3MA induced significant LC3 II enrichment in naked mole rat HSCs, which highlighted the positive effect of 3MA on autophagy. In this study, we cultured cells in serum starvation medium containing 2.5–10 mmol/L 3MA for 6–24 h. We observed that 3MA inhibited the expression of autophagy markers in a time- and dose-dependent manner. Wang et al. [37] demonstrated that 3MA attenuates CCl4-induced liver fibrosis in mice and inhibits the expression of LC3B, Beclin1, and the transcriptional regulator NF-κB in HSCs in vivo. In the present study, we investigated the effect of 3MA on IGFBPrP1-treated primary HSCs. 3MA significantly suppressed the expression of autophagy and activation markers (LC3B, Beclin1, α-SMA, and collagen I) in HSCs treated with both AdIGFBPrP1 and 3MA than those of cells treated with AdIGFBPrP1 alone. The autophagy flux and AVOs were also reduced. Furthermore, 3MA also inhibited the expression of IGFBPrP1 and TGFβ1. Thus, 3MA inhibits the effect of IGFBPrP1 on HSCs by downregulating autophagy.
To further determine whether IGFBPrP1 knockdown will block serum starvation or rapamycin induced autophagy and HSCs activation, we used shIGFBPrP1 to transfect HSCs in 2% FBS with or without rapamycin for 24 h. Previously, we observed that siRNA-mediated gene silencing of IGFBPrP1 resulted in significantly decreased levels of collagen I and fibronectin in HSCs [15]. Compared to these groups treated with or without rapamycin under serum starvation, cells transfected with shIGFBPrP1 showed a decrease in IGFBPrP1, TGFβ1, α-SMA and collagen I protein and mRNA levels; these results are similar with earlier results, which indicated that downregulation of IGFBPrP1 inhibited serum starvation or rapamycin induced HSCs activation. Furthermore, shIGFBPrP1 downregulated the expression of both LC3B and Beclin1. The autophagy flux and AVOs were also reduced. These suggested that shIGFBPrP1 blocks serum starvation or rapamycin induced autophagy.
To further understand the mechanism of IGFBPrP1-mediated autophagy and activation, we studied the PI3K/Akt/mTOR pathway, which is important in autophagy. Wu et al. [38] showed that quercetin inhibited HSCs activation and autophagy. They further found that PI3K expression was suppressed in BDL or CCl4 liver fibrosis models and increased by quercetin treatment. Akt expression was not significantly changed in fibrosis models, but p-Akt expression was increased by quercetin. Li et al. [39] found that HMGB1 showed an ability to enhance both autophagy and fibrogenesis of LX-2 cells in a time- and dose-dependent manner. Western blots data indicated HMGB1 treatment significantly decreased p-mTOR in a time-dependent manner. We observed downregulation of p-Akt and p-mTOR levels in cells treated with AdIGFBPrP1, indicating that IGFBPrP1 may promote the activation of primary HSCs by regulating the phosphorylation of Akt and mTOR, and reduce the kinase activity of mTOR to promote autophagy.
In summary, our study improves general understanding of the profibrotic mechanisms of IGFBPrP1-mediated regulation of HSCs activation partially via autophagy. However, we investigated the above-mentioned mechanism in vitro, which is the limitation of this study. Therefore, we are currently investigating the in vivo interactions between IGFBPrP1 and autophagy. In conclusion, our study revealed that IGFBPrP1 may activate HSCs and ECM expression by regulating autophagy. IGFBPrP1 may act as a potential therapeutic target for liver fibrosis. Further investigations are warranted to elucidate the in vivo mechanisms of IGFBPrP1-induced liver fibrosis.
Acknowledgments
We thank Yangyang Kong for assisting with the primary cell isolation. Lixin Liu designed the experiments. Yuzheng Zhou analyzed the data and wrote the paper. Qianqian Zhang, Xiaohong Guo, Haiyan Zhang, and Huiqin Fan performed the experiments. All authors contributed to the interpretation of the study and to further drafts. Yuzheng Zhou is the guarantor.
Author’s contribution
LLX designed the experiments. ZYZ analyzed the data and wrote the paper. ZYZ, ZQQ, KYY, ZHY, GXH, and FHQ performed the experiments. All authors contributed to the interpretation of the study and to further drafts. LLX is the guarantor.
Funding
This study was funded by the National Natural Science Foundation of China (No. 81670559).
Compliance with Ethical Standards
Conflict of interest
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
Ethical approval
All the animal protocol procedures were approved by the Shanxi Medical University Animal Care and Use Committee (SCXK2009-0001).
Footnotes
This article has been retracted. Please see the retraction notice for more detail: 10.1007/s10620-024-08506-6
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/29/2024
This article has been retracted. Please see the Retraction Notice for more detail: 10.1007/s10620-024-08506-6
References
- 1.Yin X, Yi H, Wu W, Shu J, Wu X, Yu L. R-spondin2 activates hepatic stellate cells and promotes liver fibrosis. Dig Dis Sci. 2014;59:2452–2461. doi: 10.1007/s10620-014-3208-1. [DOI] [PubMed] [Google Scholar]
- 2.Czaja MJ. Function of autophagy in nonalcoholic fatty liver disease. Dig Dis Sci. 2016;61:1304–1313. doi: 10.1007/s10620-015-4025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gual P, Gilgenkrantz H, Lotersztajn S. Autophagy in chronic liver diseases: a friend rather than a foe? Med Sci: M/S. 2017;33:252–259. doi: 10.1051/medsci/20173303011. [DOI] [PubMed] [Google Scholar]
- 4.Ueno T, Komatsu M. Autophagy in the liver: functions in health and disease. Nat Rev Gastroenterol Hepatol. 2017;14:170–184. doi: 10.1038/nrgastro.2016.185. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong W, Ju L, Qiu M, et al. Liraglutide ameliorates nonalcoholic fatty liver disease by enhancing mitochondrial architecture and promoting autophagy through the SIRT1/SIRT3-FOXO3a pathway. Hepatol Res: Off J Jpn Soc Hepatol. 2016;46:933–943. doi: 10.1111/hepr.12634. [DOI] [PubMed] [Google Scholar]
- 7.He W, Wang B, Yang J, et al. Chloroquine improved carbon tetrachloride-induced liver fibrosis through its inhibition of the activation of hepatic stellate cells: role of autophagy. Biol Pharm Bull. 2014;37:1505–1509. doi: 10.1248/bpb.b14-00297. [DOI] [PubMed] [Google Scholar]
- 8.Hernandez-Gea V, Ghiassi-Nejad Z, Rozenfeld R, et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie ZY, Xiao ZH, Wang FF. Inhibition of autophagy reverses alcohol-induced hepatic stellate cells activation through activation of Nrf2-Keap1-ARE signaling pathway. Biochimie. 2018;147:55–62. doi: 10.1016/j.biochi.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 10.Jiang JX, Torok NJ. Liver injury and the activation of the hepatic myofibroblasts. Curr Pathobiol Rep. 2013;1:215–223. doi: 10.1007/s40139-013-0019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukada H, Yamashina S, Izumi K, et al. Suppression of autophagy sensitizes Kupffer cells to endotoxin. Hepatol Res: Off J Jpn Soc Hepatol. 2012;42:1112–1118. doi: 10.1111/j.1872-034X.2012.01024.x. [DOI] [PubMed] [Google Scholar]
- 12.Weiskirchen R, Tacke F. Cellular and molecular functions of hepatic stellate cells in inflammatory responses and liver immunology. Hepatobiliary Surg Nutr. 2014;3:344–363. doi: 10.3978/j.issn.2304-3881.2014.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao CF. Transforming growth factor beta 1 and liver cirrhosis. Zhonghua yi xue za zhi. 2005;85:1074–1077. [PubMed] [Google Scholar]
- 14.Guo Y, Zhang Y, Zhang Q, et al. Insulin-like growth factor binding protein-related protein 1 (IGFBPrP1) contributes to liver inflammation and fibrosis via activation of the ERK1/2 pathway. Hepatol Int. 2015;9:130–141. doi: 10.1007/s12072-014-9578-9. [DOI] [PubMed] [Google Scholar]
- 15.Guo XH, Liu LX, Zhang HY, et al. Insulin-like growth factor binding protein-related protein 1 contributes to hepatic fibrogenesis. J Dig Dis. 2014;15:202–210. doi: 10.1111/1751-2980.12126. [DOI] [PubMed] [Google Scholar]
- 16.Li XQ, Zhang QQ, Zhang HY, Guo XH, Fan HQ, Liu LX. Interaction between insulin-like growth factor binding protein-related protein 1 and transforming growth factor beta 1 in primary hepatic stellate cells. Hepatobiliary Pancreat Dis Int. 2017;16:395–404. doi: 10.1016/S1499-3872(17)60013-4. [DOI] [PubMed] [Google Scholar]
- 17.Thomes PG, Brandon-Warner E, Li T, Donohue TM, Jr, Schrum LW. Data on the effect of pro-fibrotic cytokine TGF-beta on hepatic stellate cell autophagy. Data Brief. 2016;10:312–314. doi: 10.1016/j.dib.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu MY, He YJ, Lv X, et al. Transforming growth factorbeta1 reduces apoptosis via autophagy activation in hepatic stellate cells. Mol Med Rep. 2014;10:1282–1288. doi: 10.3892/mmr.2014.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paquette M, El-Houjeiri L, Pause A. mTOR pathways in cancer and autophagy. Cancers. 2018;10:18. doi: 10.3390/cancers10010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamanaka Y, Wilson EM, Rosenfeld RG, Oh Y. Inhibition of insulin receptor activation by insulin-like growth factor binding proteins. J Biol Chem. 1997;272:30729–30734. doi: 10.1074/jbc.272.49.30729. [DOI] [PubMed] [Google Scholar]
- 21.Evdokimova V, Tognon CE, Benatar T, et al. IGFBP7 binds to the IGF-1 receptor and blocks its activation by insulin-like growth factors. Sci Sig. 2012;5:ra92. doi: 10.1126/scisignal.2003184. [DOI] [PubMed] [Google Scholar]
- 22.Feng J, Niu P, Chen K, et al. Salidroside mediates apoptosis and autophagy inhibition in concanavalin A-induced liver injury. Exp Ther Med. 2018;15:4599–4614. doi: 10.3892/etm.2018.6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo T, Liu G, Ma H, et al. Inhibition of autophagy via activation of PI3K/Akt pathway contributes to the protection of ginsenoside Rb1 against neuronal death caused by ischemic insults. Int J Mol Sci. 2014;15:15426–15442. doi: 10.3390/ijms150915426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Zhang QQ, Guo XH, Zhang HY, Liu LX. IGFBPrP1 induces liver fibrosis by inducing hepatic stellate cell activation and hepatocyte apoptosis via Smad2/3 signaling. World J Gastroenterol. 2014;20:6523–6533. doi: 10.3748/wjg.v20.i21.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo X, Zhang H, Zhang Q, Li X, Liu L. Screening for and validation of a hepatic fibrosis-related pathway induced by insulin-like growth factor-binding protein-related protein 1. Eur J Gastroenterol Hepatol. 2016;28:762–772. doi: 10.1097/MEG.0000000000000631. [DOI] [PubMed] [Google Scholar]
- 26.Decaris ML, Li KW, Emson CL, et al. Identifying nonalcoholic fatty liver disease patients with active fibrosis by measuring extracellular matrix remodeling rates in tissue and blood. Hepatology (Baltimore, Md.) 2017;65:78–88. doi: 10.1002/hep.28860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan Y, Zhang Y, Han X, et al. Relaxin alleviates TGFbeta1-induced cardiac fibrosis via inhibition of Stat3-dependent autophagy. Biochem Biophys Res Commun. 2017;493:1601–1607. doi: 10.1016/j.bbrc.2017.09.110. [DOI] [PubMed] [Google Scholar]
- 28.Ni BB, Li B, Yang YH, et al. The effect of transforming growth factor beta1 on the crosstalk between autophagy and apoptosis in the annulus fibrosus cells under serum deprivation. Cytokine. 2014;70:87–96. doi: 10.1016/j.cyto.2014.07.249. [DOI] [PubMed] [Google Scholar]
- 29.Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol Biol (Clifton, N.J.) 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- 30.Boyle KB, Randow F. The role of ‘eat-me’ signals and autophagy cargo receptors in innate immunity. Curr Opin Microbiol. 2013;16:339–348. doi: 10.1016/j.mib.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 31.Sahani MH, Itakura E, Mizushima N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy. 2014;10:431–441. doi: 10.4161/auto.27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sotthibundhu A, McDonagh K, von Kriegsheim A, et al. Rapamycin regulates autophagy and cell adhesion in induced pluripotent stem cells. Stem cell Res Ther. 2016;7:166. doi: 10.1186/s13287-016-0425-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen M, Liu J, Yang L, Ling W. AMP-activated protein kinase regulates lipid metabolism and the fibrotic phenotype of hepatic stellate cells through inhibition of autophagy. FEBS Open Bio. 2017;7:811–820. doi: 10.1002/2211-5463.12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y, Wang W, Peng XM, et al. Rapamycin upregulates connective tissue growth factor expression in hepatic progenitor cells through TGF-beta-Smad2 dependent signaling. Front Pharmacol. 2018;9:877. doi: 10.3389/fphar.2018.00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu YT, Tan HL, Shui G, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao S, Lin L, Kan G, et al. High autophagy in the naked mole rat may play a significant role in maintaining good health. Cell Physiol Biochem: Int J Exp Cell Physiol Biochem Pharmacol. 2014;33:321–332. doi: 10.1159/000356672. [DOI] [PubMed] [Google Scholar]
- 37.Wang B, Yang H, Fan Y, et al. 3-Methyladenine ameliorates liver fibrosis through autophagy regulated by the NF-kappaB signaling pathways on hepatic stellate cell. Oncotarget. 2017;8:107603–107611. doi: 10.18632/oncotarget.22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu L, Zhang Q, Mo W, et al. Quercetin prevents hepatic fibrosis by inhibiting hepatic stellate cell activation and reducing autophagy via the TGF-beta1/Smads and PI3K/Akt pathways. Sci Rep. 2017;7:9289. doi: 10.1038/s41598-017-09673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Zeng C, Zheng B, et al. HMGB1-induced autophagy facilitates hepatic stellate cells activation: a new pathway in liver fibrosis. Clin Sci (London, England: 1979) 1979;2018(132):1645–1667. doi: 10.1042/CS20180177. [DOI] [PubMed] [Google Scholar]