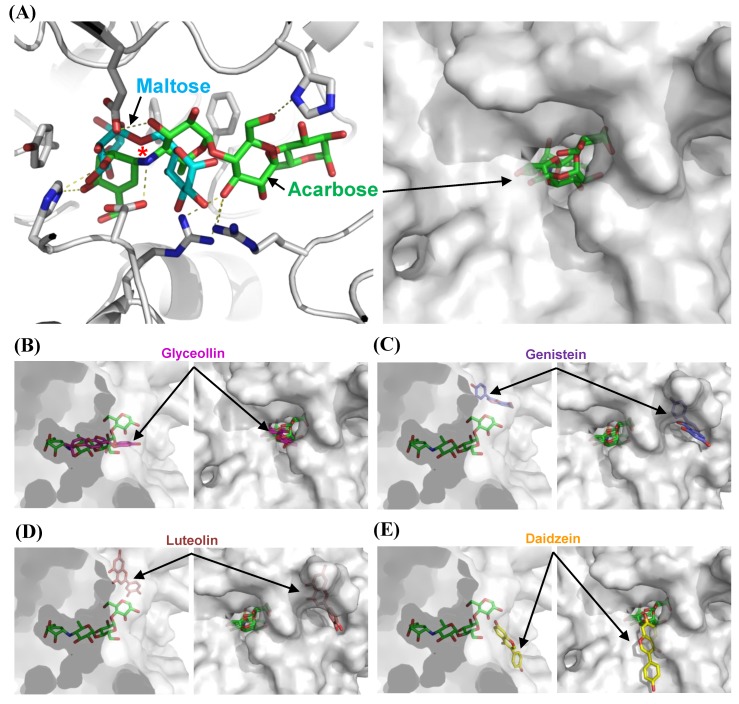
Figure 4.
Molecular docking studies on the modes of α-glucosidase inhibition. The model of MAL12, a structure of α-glucosidase, is shown as a cartoon or surface diagram in white. Isomaltose, acarbose, glyceollin, genistein, luteolin, and daidzein are shown as stick diagrams. (A) The inhibitory mode of acarbose. The isomaltose molecule was prepared by superposing the model of MAL12 with the structure of isomaltase (PDB code 3AXH) and is shown in cyan. Residues involved in substrate binding are displayed as stick models. Yellow dashed lines indicate hydrogen bond interactions. The glucosidic bonds to be hydrolyzed are indicated by a star. (B–E) The inhibitory modes of polyphenols. Glyceollin, genistein, luteolin, and daidzein are shown as stick diagrams in magenta, purple, yellow, and brown, respectively.